Sepharose High Performance: Purification with High Resolution
Use Sepharose High Performance media for purification of proteins, peptides or oligonucleotides.
Use Sepharose High Performance for intermediate purification steps that require high capacity and high resolution (flows up to 150 cm/h).
Run Sepharose High Performance columns on systems such as ÄKTAdesign, FPLC System and HPLC. Appendix 4 gives guidance on how to select the most suitable ÄKTAdesign system.
Sepharose High Performance media are based on a matrix of 34 μm particles made from 6% agarose and highly cross-linked for chemical and physical stability. The small particle size ensures fast binding and dissociation even at high sample loads and flow rates which, in combination with high selectivity, give high resolution separations. Particle size and bed volumes remain stable, despite changes in ionic strength or pH, to ensure fast separations at high flow rates. The strong ion exchange groups (Q and S) maintain their charge over a broad pH range, allowing selection of the most suitable pH for each application.
Sepharose High Performance media, as other ion exchangers, can be used for group separations or sample concentration. However, these separations should be limited to reasonably clean samples to avoid the risk of blocking the column filter (34 μm particle size requires the use of finer column filters).
Purification options

Figure 49.Q and S Sepharose High Performance media are available prepacked in HiTrap and HiLoad columns or in media packs.
*See Appendix 5 to convert linear flow (cm/hour) to volumetric flow rates (ml/min) and vice versa.
**Working pH range refers to the pH interval where the medium binds protein as intended or as needed for elution without adverse long term effects.
***Maximum operating back pressure refers to the pressure above which the medium begins to compress.
Use prepacked HiTrap columns (1 ml or 5 ml) for media selection, method scouting, group separations, small scale purification, sample concentration or clean-up. Connect up to 3 HiTrap columns in series to scale-up.
Use prepacked HiLoad columns (20 ml or 53 ml) for method development, group separations, larger scale purification or sample concentration.
For column packing:
Select a production column such as FineLINE for larger volumes.
Purification examples
Intermediate purification

Figure 50.Intermediate step from a purification protocol for a2-macroglobulin. An affinity step on Blue Sepharose was used to remove albumin, the major contaminant, before IEX.
Scaling-up
Using prepacked Sepharose High Performance columns facilitates quick scale-up and ensures reproducibility, as shown in Figures 51 and 52.
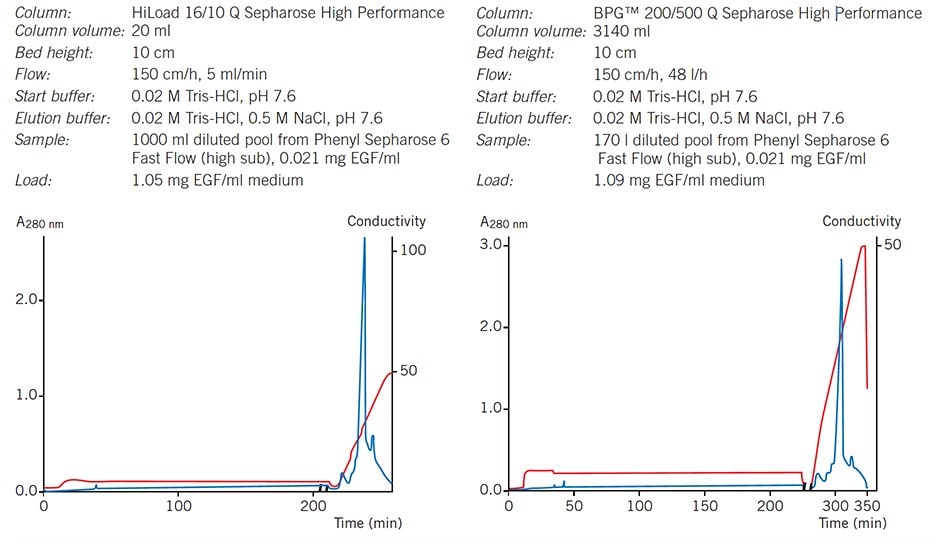
Figure 51.Elution pattern, purity and yield are maintained when scaling-up from a HiLoad column to a BPG column.

Figure 52.Reproducibility maintained as purification is scaled up from a 1 ml HiTrap column through to a 53 ml HiLoad 26/10 SP Sepharose High Performance column.
Group separations
Figure 53 shows a group separation of human serum proteins on HiTrap Q HP using a one step elution that had been optimized to ensure that IgG flowed through the column leaving other serum components to be eluted separately. Figure 54 illustrates a multi-step elution to separate groups of human milk proteins.

Figure 53.Separation of IgG from human serum proteins on HiTrap Q HP, 1 ml, using one step elution. Analysis by SDS-PAGE (Phast System, PhastGel 10–15, silver staining).
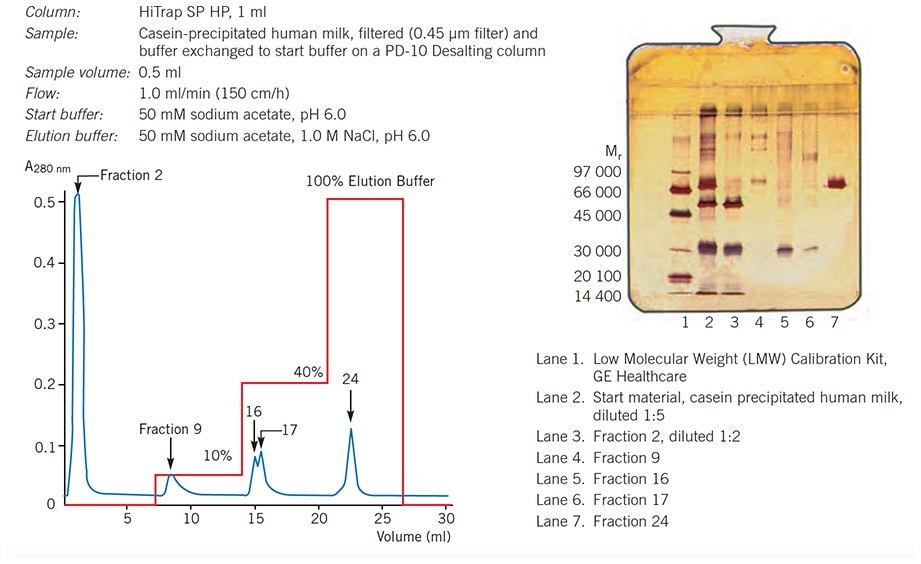
Figure 54.Separation of human milk proteins on HiTrap SP HP, 1 ml, using one step elution. Analysis by SDS-PAGE (Phast System, PhastGel 10–15, silver staining).
Sample concentration
It can be an advantage to concentrate a sample prior to gel filtration in order to minimize sample volume and facilitate a rapid, high resolution size separation. HiTrap columns offer a convenient, ready to use solution for sample concentration. Table 7 gives examples of the high concentration factors achieved when concentrating proteins from very dilute starting material using HiTrap columns prepacked with Sepharose HP medium. Similar results can be achieved with HiTrap columns prepacked with Sepharose Fast Flow or Sepharose XL media
Performing a separation
Guidelines for selection of media, buffer, pH and ionic strength conditions and method optimization are given in Chapter 2. Use the instructions given here as a basis from which to optimize a separation.
Correct sample and buffer preparation is essential in order to achieve optimal separation and avoid any deterioration in column performance. Samples must be fully dissolved and free from particles or other material likely to interfere with the separation. Refer to Chapter 2 and Appendix 1 for recommendations and advice on sample preparation.
Filter buffers after all salts and additives have been included. Use high quality water and chemicals. Filter solutions through 0.45 μm or 0.22 μm filters. To avoid formation of air bubbles in a packed column, ensure that column and buffers are at the same temperature when preparing for a run.
The pH of the start buffer should be at least 0.5–1 pH unit above the pI of the target substance when using an anion exchanger (Q) and 0.5–1 pH unit below the pI of the target substance when using a cation exchanger (S). See Appendix 2 for recommendations on volatile and non-volatile buffer systems for anion and cation exchangers.
For samples with unknown charge properties, try the following:
- anion exchange (Q)
start buffer: pH 8.0
elution buffer: start buffer including 1 M NaCl, pH 8.0 - cation exchange (S)
start buffer: pH 6.0
elution buffer: start buffer including 1 M NaCl, pH 6.0
Users of ÄKTAdesign systems with BufferPrep functionality can select one of the buffer recipes recommended for anion exchange chromatography at pH 8 or cation exchange chromatography at pH 6.
First time use or after long term storage:
- To remove ethanol, wash with 1 column volume of distilled water at 1 ml/min (HiTrap 1 ml), 5 ml/min (HiTrap 5 ml), 0.8 ml/min (HiLoad 20 ml), 2.2 ml/min (HiLoad 53 ml) or at 25 cm/h for Sepharose High Performance packed in larger columns. This step ensures removal of ethanol and avoids the risk of precipitation if buffer salts were to come into contact with the ethanol. The step can be omitted if precipitation is not likely to be a problem.
- Wash with 5 column volumes of start buffer, at 1 ml/min (HiTrap 1 ml), 5 ml/min (HiTrap 5 ml), 3 ml/min (HiLoad 20 ml), 8 ml/min (HiLoad 53 ml) or at 50 cm/h for Sepharose High Performance packed in larger columns.
- Wash with 5 column volumes of elution buffer, same flow as step 2.
- Wash with 5 column volumes of start buffer, same flow as step 2.
- Run a blank elution before applying sample.
Separation by gradient elution
Flow: 1 ml/min (HiTrap 1 ml), 5 ml/min (HiTrap 5 ml), 3 ml/min (HiLoad 20 ml), 8 ml/min (HiLoad 53 ml) or at 50–100 cm/h for Sepharose High Performance packed in larger columns. Collect fractions throughout the separation.
- Equilibrate column with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable.
- Adjust the sample to the chosen starting pH and ionic strength and apply to the column.
- Wash with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable i.e. when all unbound material has washed through the column.
- Begin elution using a gradient volume of 10–20 column volumes and an increasing ionic strength up to 0.5 M NaCl (50%B).
- Wash with 5 column volumes of 1 M NaCl (100%B) to elute any remaining ionically-bound material.
- Re-equilibrate with 5–10 column volumes of start buffer or until eluent pH and conductivity reach the required values.
Separation by step elution
Flow: 1 ml/min (HiTrap 1 ml), 5 ml/min (HiTrap 5 ml), 3 ml/min (HiLoad 20 ml), 8 ml/min (HiLoad 53 ml) or at 50–100 cm/h for Sepharose High Performance packed in larger columns. Collect fractions throughout the separation.
- Equilibrate column with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable.
- Adjust the sample to the chosen starting pH and ionic strength and apply to the column.
- Wash with 5–10 column volumes of start buffer or until the baseline, eluent pH and conductivity are stable i.e. when all unbound material has washed through the column.
- Elute with 5 column volumes of start buffer + NaCl at chosen ionic strength.
- Repeat step 4 at higher ionic strengths until the target protein(s) has been eluted.
- Wash with 5 column volumes of a high salt solution (1 M NaCl in start buffer) to elute any remaining ionically bound material.
- Re-equilibrate with 5–10 column volumes of start buffer or until eluent pH and conductivity reach the required values.
Save time by using higher flow rates during the high salt wash and re-equilibration steps. Do not exceed the maximum recommended flow for the medium.
If ionic detergents have been used, wash the column with 5 column volumes of distilled water, followed by 2 column volumes 2 M NaCl. Re-equilibrate with at least 10 column volumes of start buffer until the UV baseline, eluent pH and/or conductivity are stable. Organic solvents such as ethanol can be used to remove non-ionic detergents. When selecting an organic solvent, check the chemical stability of the medium to determine a suitable concentration.
Check column performance regularly by determining column efficiency and peak symmetry. See Appendix 3. Note that this does not apply to HiTrap columns.
Cleaning
Correct preparation of samples and buffers and application of a high salt wash (1 M NaCl) at the end of each separation should keep most columns in good condition. However, reduced performance, a slow flow rate, increasing back pressure or complete blockage are all indications that the medium needs to be cleaned using more stringent procedures in order to remove contaminants.
It is recommended to reverse the direction of flow during column cleaning so that contaminants do not need to pass through the entire length of the column. The number of column volumes and time required for each cleaning step may vary according to the degree of contamination. If the cleaning procedure to remove common contaminants does not restore column performance, change the top filter (when possible) before trying alternative cleaning methods. Care should be taken when changing a filter as this may affect the column packing and interfere with performance.
The following procedure should be satisfactory to remove common contaminants:
- Wash with at least 2 column volumes of 2 M NaCl at 1 ml/min (HiTrap 1 ml), 5 ml/min (HiTrap 5 ml), 3 ml/min (HiLoad 20 ml), 8 ml/min (HiLoad 53 ml) or at 40 cm/h with a contact time of 1–2 hours for Sepharose High Performance packed in larger columns.
- Wash with at least 4 column volumes of 1 M NaOH (same flow as step 1).
- Wash with at least 2 column volumes of 2 M NaCl (same flow as step 1).
- Rinse with at least 2 column volumes of distilled water (same flow as step 1) until the UV-baseline and the eluent pH are stable.
- Wash with at least 4 column volumes of start buffer or storage buffer (same flow as step 1) until eluent pH and conductivity have reached the required values.
To remove precipitated proteins, lipids, hydrophobically bound proteins or lipoproteins, refer to Appendix 1.
Media characteristics
Composition: sulfopropyl (SP) or quaternary amino (Q) groups coupled to highly cross-linked 6% agarose via chemically stable ether bonds.
*Long term pH stability refers to the pH interval where the medium is stable over a long period of time without adverse side effects on the chromatography performance.
Short term pH stability refers to the pH interval for regeneration, cleaning-in-place and sanitization procedures.
All ranges are estimates based on the experience and knowledge within Cytiva.
Chemical stability
For daily use, Sepharose High Performance media are stable in all common, aqueous buffers, 1 M NaOH, denaturing agents (8 M urea, 6 M guanidine hydrochloride), 70% ethanol, 1 M acetic acid, 30% acetonitrile and with additives such as non-ionic detergents. Sepharose High Performance can be used with organic solvents such as dimethylsulfoxide, dimethylformamide, tetrahydrofuran, acetone, chloroform, dichloromethane, dichloroethane and dichloroethane/pyridine (50:50) as well as polar solvents and aqueous/organic isolutions. The water in the medium can be exchanged by the alternative solvent with very little effect on the pore size of the matrix.
Avoid cationic detergents with SP Sepharose High Performance. Avoid anionic detergents with Q Sepharose High Performance. Avoid oxidizing agents.
Storage
For column storage, wash with 2 column volumes of distilled water followed by 2 column volumes of 20% ethanol. Include 0.2 M sodium acetate in the 20% ethanol solution for columns packed with SP Sepharose High Performance. Degas the ethanol/water mixture thoroughly and apply at a low flow rate to avoid over-pressuring the column. Store at room temperature or, for long periods, store at +4 °C to +8 °C. Ensure that the column is sealed well to avoid drying out. Whenever possible, use the storage and shipping device if supplied by the manufacturer. Store unused media at +4 °C to +30 °C in 20% ethanol. Do not freeze.
To avoid formation of air bubbles in a packed column, ensure that column and buffers are at the same temperature when preparing for a run.
Materials
To continue reading please sign in or create an account.
Don't Have An Account?