Proteinase K in Tissue Samples
Read more about
- What is the Role of Proteinase K in Isolating Nucleic Acids in Tissue Samples?
- How Do You Isolate Nucleic Acids From Tissue Samples With Proteinase K
- How Do You Isolate Nucleic Acids With Proteinase K From Ffpe Tissue Samples?
- Other Proteinase K Information
- How to Order Large Quantities of Proteinase K
- Materials
- References
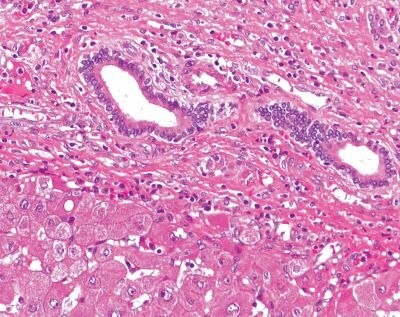
Hematoxylin and eosin (H&E) stained formalin-fixed, paraffin-embedded (FFPE) liver tissue
Proteinase K is a broad-spectrum serine protease that is commonly used in the isolation of nucleic acids from tissue samples. Its role is to digest proteins in the tissue sample, which can interfere with the isolation and purification of nucleic acids.
Tissue samples usually contain various proteins, including histones, nucleases, and other enzymes, that can degrade DNA and RNA, and interfere with downstream applications such as PCR and sequencing. Proteinase K can break down these proteins, releasing the nucleic acids from the tissue matrix and reducing their degradation by endogenous enzymes.
Proteinase K can efficiently digest proteins in a wide range of conditions, including high temperatures, high salt concentrations, and the presence of detergents. It is also stable at high temperatures, making it suitable for heat inactivation after the digestion step.
After the digestion with proteinase K, the nucleic acids can be further purified using various methods such as phenol-chloroform extraction, column-based purification, or magnetic bead-based purification. The resulting purified nucleic acids can then be used for downstream applications such as PCR, sequencing, or hybridization assays.
How do you isolate nucleic acids from tissue samples with proteinase K?
The isolation of nucleic acids from tissue samples typically involves several steps, including tissue homogenization, protein digestion with proteinase K, and purification of the nucleic acids. Here is a general overview of the procedure:
1. Tissue homogenization: The tissue sample is first homogenized to disrupt the tissue matrix and release the nucleic acids. This can be done using a mechanical homogenizer, a blender, or a bead mill.
2. Protein digestion: Proteinase K is added to the homogenized tissue sample to digest the proteins and release the nucleic acids. The amount of proteinase K required can vary depending on the tissue type and the amount of tissue used. Typically, 100-200 μg of proteinase K per mL of tissue homogenate is used. Digestion can be performed at 37-65 °C for 30 minutes to several hours, depending on the tissue type and the extent of protein digestion required.
3. Inactivation of proteinase K: After protein digestion, the proteinase K is typically inactivated by heating the sample at 95-100 °C for 10-15 minutes. This helps to ensure that the proteinase K is fully inactivated and does not interfere with downstream applications.
4. Nucleic acid purification: The nucleic acids are then purified from the proteinase K-treated sample. This can be done using various methods, including phenol-chloroform extraction, column-based purification, or magnetic bead-based purification. The choice of purification method will depend on factors such as the type and amount of nucleic acid required, the downstream application, and the available equipment.
After purification, the nucleic acids can be quantified using a spectrophotometer or a fluorometer, and their quality can be assessed using gel electrophoresis or other methods. The purified nucleic acids can then be used for downstream applications such as PCR, sequencing, or hybridization assays.
Proteinase K can help in the purification of DNA from FFPE tissue samples by degrading the cross-linked proteins that are bound to the DNA.
How do you isolate nucleic acids with proteinase K from FFPE tissue samples?
Isolation of nucleic acids from formalin-fixed paraffin-embedded (FFPE) tissue samples can be challenging due to the cross-linking of proteins and nucleic acids caused by the formalin fixation process. However, proteinase K can be used to digest the protein cross-links and release the nucleic acids from the tissue. Here is a general overview of the procedure for isolating nucleic acids from FFPE tissue samples with proteinase K:
1. Deparaffinization: The FFPE tissue sample is first deparaffinized to remove the paraffin wax and expose the tissue for subsequent steps. This can be done using xylene or other organic solvents.
2. Rehydration: The deparaffinized tissue is then rehydrated by washing in a series of graded alcohols (e.g., 100% ethanol, 95% ethanol, 70% ethanol, and water).
3. Protein digestion: Proteinase K is added to the rehydrated tissue to digest the proteins and release the nucleic acids. The amount of proteinase K required can vary depending on the tissue type and the amount of tissue used. Typically, 100-200 μg of proteinase K per mL of tissue homogenate is used. Digestion can be performed at 37-65 °C for several hours to overnight, depending on the extent of protein digestion required.
4. Inactivation of proteinase K: After protein digestion, the proteinase K is typically inactivated by heating the sample at 95-100 °C for 10-15 minutes. This helps to ensure that the proteinase K is fully inactivated and does not interfere with downstream applications.
5. Nucleic acid purification: The nucleic acids are then purified from the proteinase K-treated sample. This can be done using various methods, including phenol-chloroform extraction, column-based purification, or magnetic bead-based purification. The choice of purification method will depend on factors such as the type and amount of nucleic acid required, the downstream application, and the available equipment.
After purification, the nucleic acids can be quantified using a spectrophotometer or a fluorometer, and their quality can be assessed using gel electrophoresis or other methods. The purified nucleic acids can then be used for downstream applications such as PCR, sequencing, or hybridization assays.
There are several variations of this protocol, and the specific details can vary depending on the type of nucleic acid being isolated and the downstream application. However, the use of proteinase K digestion is a common step in many protocols for isolating nucleic acids from FFPE tissue samples.
Other Proteinase K Information
To learn how Proteinase K is used blood DNA/RNA extraction, visit How Proteinase K Can Be Used in Blood DNA Extraction.
To learn different Proteinase K protocols here visit Guide on How to Use Proteinase K in Different Procedures.
How do you find the best Proteinase K supplier, visit How to Determine the Best Product and Supplier for Proteinase K.
How to Order Large Quantities of Proteinase K
Bulk capabilities and custom pack sizes of Proteinase K are available for your specific application. Complete the form to receive more information on what we have to offer.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?