p53: DNA Damage Response and Cancer
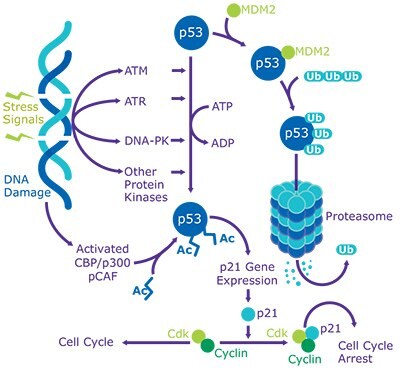
p53 is a transcription factor that binds to defined consensus sites within DNA. A major function of p53 is to repress genes involved in cell growth stimulation while activating alternate genes involved in cell cycle control. p53 regulates gene expression to control a wide range of biological activities that allow rapid cell adaptation to environmental changes. The most prominent role of p53 is in growth arrest before either DNA replication in the G1 phase or before mitosis in the G2 phase. This provides a window for DNA repair or elimination of cells with severely damaged DNA. p53 activation can result in apoptosis and serve as a means to eliminate severely damaged cells by transcriptional activation of pro-apoptotic genes or through transcription-independent mechanisms.
p53 signaling is activated in cells as a response to various signals during carcinogenesis. Carcinogen-induced DNA damage, abnormal proliferative signals, hypoxia, and loss of cell adhesion are some of the most common signals that activate p53. The final outcome of p53 activation depends on many factors, and is mediated largely through the action of downstream effector genes transactivated by p53. Agents that damage DNA induce p53 to become very stable by post-translational mechanisms, allowing p53 levels in the nucleus to increase dramatically. Hence, p53 can suppress tumors by ensuring genomic integrity and repressing proliferation of tumor-forming cells.
Regulation of p53 transcriptional activity by MDM2
In unstressed cells, p53 is latent and maintained at low levels by targeted, ubiquitin-mediated degradation by MDM2/HDM2 and many other ubiquitin ligases. MDM2 is a p53-inducible phosphoprotein that binds to the N-terminus of p53 and negatively regulates its activity. Interestingly, transcription of Mdm2 is activated by p53. Hence, in the presence of high levels of p53, MDM2 levels are also elevated. p53 interacts with MDM2 at Phe19, Trp23, and Leu26 to fill up a complementary hydrophobic pocket of MDM2. These three amino acids are also essential for transactivation of p53. Binding of MDM2 to p53 antagonizes the transcriptional activity of p53 and blocks its acetylation and transactivation by interfering with transcriptional co-activators, p300/CBP. MDM2 functions as an E3 ligase to ubiquitinate p53 and force its export from the nucleus to the cytoplasm, where p53 is degraded by the proteasome.
Site-specific phosphorylation and acetylation of p53
Human p53 is phosphorylated at multiple sites by stress-activated protein kinases, DNA-dependent protein kinase (DNA-PK), casein kinase I and II, and cyclin-dependent kinases. Most of the p53 phosphorylation sites are clustered within the N-terminal transactivation domain. Phosphorylation of p53 provides stability by promoting its dissociation from MDM2 and enhancing its transcriptional activity. Additionally, ATM and ATR kinases promote phosphorylation of human p53 at Ser15 and Ser20, which are essential for the activation of p53 following DNA damage. DNA-PK phosphorylates Ser15 within the critical N-terminal region of p53, which controls the interaction of p53 with the transcriptional apparatus and with MDM2. DNA-PK also phosphorylates Ser9 and Thr18; however, phosphorylation at these sites is dependent upon the presence of the full-length p53, and is independent of phosphorylation at other sites. When p53 co-localizes with DNA-PK and ssDNA, there is a 10-fold enhancement of p53 phosphorylation. All types of tumor cells are shown to exhibit higher levels of p53 phosphorylation when compared to normal non-transformed cells, which offer greater stability to p53 regardless of any p53 mutations. Despite extensive studies on p53 phosphorylation, it is now known that phosphorylation is not the only mechanism that regulates p53 activation. Following cellular stress, p53 is shown to be acetylated by p300/CBP at multiple lysine residues (Lys370, 372, 373, 381, and 382) and by pCAF at Lys320. In vitro studies show that increasing the level of p53 acetylation with deacetylase inhibitors prevents the degradation of p53. HDAC1 and SIRT1 are among the deacetylases that may be responsible for regulating p53.
p53 and cancer
p53 has been shown to be either non-functional or mutated in most human cancers. A large number of mutations are caused by single base substitutions, and about 30% of these mutations are reported to occur in hotspot codons. Functional p53 provides a protective mechanism against tumor growth, and a loss of p53 function is a key step in the neoplastic cascade. A successful response to therapy is greatly reduced in tumors where mutant p53 is present, and these tumors are often very difficult to treat. Future genome-wide studies and the increase in the use of an “omic” approach are likely necessary to elucidate the diverse mechanisms of action for p53 and its role in regulating gene expression and cancer.
Materials
To continue reading please sign in or create an account.
Don't Have An Account?