Optimizing Crude Plant Samples for PCR
Overcoming Potent Inhibitors and sample diversity in direct plant PCR
Success rates with direct PCR are typically low due to the high concentrations of inhibitors present in even small amounts of plant material, and the precise control of sample size required. In this Application Note, a systematic approach for reaction optimization with the KAPA3G Plant PCR Kit is presented, as well as a robust method for successful crude sample PCR.
PCR From Purified DNA
Annealing temperature optimization was performed with two universal primer sets, Tab a/b and Tab c/d, targeting the trnL intron (Taberlet et al.). Genomic DNA purified from Erica cerinthoides and Cinnamomum camphora was used as template. Results (Figure 1) showed that the Tab a/b primer set worked best at an annealing temperature of 56° C, while the Tab c/d primer set worked well across the entire annealing temperature range tested. The Tab c/d primer set was selected for this study, using an annealing temperature of 55° C. In the next experiment (Figure 2), the Tab c/d primer set was used with genomic DNA purified from 18 different plant sample types from various species. Amplification was successful for all 18 plants tested.
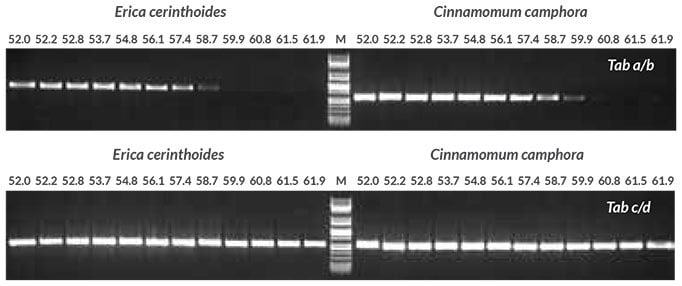
Figure 1.Annealing temperature optimization with purified DNA Annealing temperature gradient PCR (52 to 62° C) was performed to determine the optimal annealing temperature for two primer sets targeting the trnL intron, Tab a/b and Tab c/d (Taberlet et al.). Reactions (50 μL) contained 1X KAPA Plant PCR Buffer (providing 1.5 mM MgCl2 and 0.2 mM of each dNTP), 0.3 μM of each primer, 1 U of KAPA3G Plant DNA Polymerase and 1 μL of purified DNA extracted from Erica cerinthoides and Cinnamomum camphora using the QIAGEN® DNeasy Plant Mini Kit (10 ng per reaction).
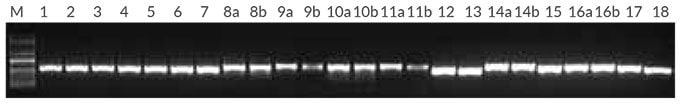
Figure 2.Plant PCR with purified DNA from 18 sample types Reactions (50 μL) contained 1X KAPA Plant PCR Buffer (providing 1.5 mM MgCl2 and 0.2 mM of each dNTP), 0.3 μM of each primer for the Tab c/d primer set, targeting the trnL intron (Taberlet et al.), 1 U of KAPA3G Plant DNA Polymerase and 1 μL of purified DNA extracted using the Qiagen DNeasy Plant Mini Kit (1 – 10 ng per reaction). Plants used were 1 – 7: Erica spp. (leaves); 8 – 12: Euphorbia spp. (leaves [a] and thorns [b]); 13: Euryops tysonii (leaf); 14: Schotia brachypetala (leaf [a] and seedpod [b]); 15: Hyaenanche globosa (leaf); 16: Acacia sieberiana (leaf [a] and seedpod [b]); 17: Cinnamomum camphora (leaf); 18: Fragaria ananassa (leaf). A standard 35-cycle plant PCR was performed, with an annealing temperature of 55° C, and 15 sec extension at 72° C.
Direct PCR
The success rate of direct PCR with eight plant species, including several crops, was evaluated using either a 0.35 mm or 0.5 mm piece of leaf/seed. Results are shown in Figure 3. Generally, leaf samples amplified well, with the exception of grapevine (Vitis vinifera) and Eucalyptus spp. Seeds had a lower success rate, and tended to result in nonspecific amplification.
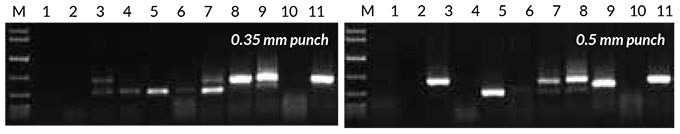
Figure 3. Direct PCR with the Tab c/d primer pair was performed on leaves and/or seeds from eight plants, sampled using either a 0.35 mm punch (left) or 0.5 mm punch (right). Reactions were set up as described in the KAPA3G Plant PCR Kit Technical Data Sheet, and 45 cycles of PCR performed with annealing at 55° C, and 20 sec extension at 72° C per cycle.
1: Eucalyptus; 2: Grapevine; 3: Wheat leaf; 4: Wheat seed; 5: Canola leaf; 6: Canola seed; 7: Rice seed; 8: Barley seed; 9: Corn kernel; 10: Cotton seed; 11: Cotton leaf.
Crude Sample Extraction and PCR
Crude extracts were prepared from leaves/seeds of the same eight plant species used for direct PCR, as described in the KAPA3G Plant PCR Kit Technical Data Sheet—both with and without heat treatment (5 min at 95°C). Of this crude extract, 1 μL was used as template in the PCR reaction. Results (Figure 4) show that all samples amplified successfully. The heat treatment increased yields slightly for all of the plants with the exception of Eucalyptus.
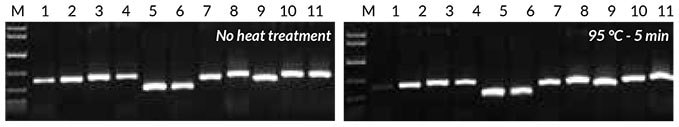
Figure 4.Results obtained with crude sample PCR Crude sample PCR with the Tab c/d primer pair was performed on leaves and/ or seeds from eight plants, with 1 μL of crude extract prepared without heat treatment (left) or 1 μL of crude extract prepared with heat treatment (right). Reactions were set up as described in the KAPA3G Plant PCR Kit Technical Data Sheet, and 45 cycles of PCR performed with annealing at 55° C, and 20 sec extension at 72° C per cycle.
1: Eucalyptus; 2: Grapevine; 3: Wheat leaf; 4: Wheat seed; 5: Canola leaf; 6: Canola seed; 7: Rice seed; 8: Barley seed; 9: Corn kernel; 10: Cotton seed; 11: Cotton leaf.
Optimization of Crude Sample Plant PCR
While success rates with crude sample PCR are typically high, there are exceptions where optimization is required in addition to the initial optimization with purified DNA. Reaction parameters to be optimized include the enzyme and MgCl2 concentrations, and the use of an optional additive (e.g., 1 mM TCEP). Cycling parameters generally do not require much optimization, but in certain cases, the cycle number and extension time may have to be increased. Re-optimization of the annealing temperature may also be necessary.
To demonstrate the optimization process, crude extracts prepared from linseed were amplified with eight combinations of enzyme (1 or 2 U), MgCl2 (2 or 3 mM) and TCEP (0 or 1 mM). Refer to Table 1 for full details. This was combined with annealing temperature gradient PCR at each condition, to determine whether the temperature optimized initially remained optimal. Two of the reaction conditions (F and G, highlighted in red in Table 1) were shown to improve the yield of product obtained, while an annealing temperature of 55°C remained suitable for this assay. Improved results were obtained with 2 U of enzyme and 3 mM MgCl2 (panel G), or with 2 U of enzyme, 2 mM MgCl2 and 1 mM TCEP (panel F). Should results not have improved significantly with the combinations tested, the cycle number and extension time would have been increased in future rounds of optimization.
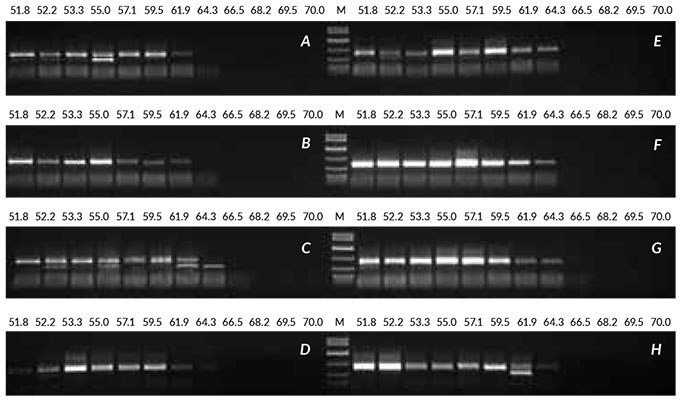
Figure 5. Results obtained with optimization of crude sample from linseed Crude sample PCR from linseed was optimized by performing eight sets of annealing temperature gradient PCRs (52 to 70° C), all containing 1 μL of crude extract (prepared without heat treatment). Reactions were performed with various combinations of enzyme, MgCl2 and TCEP, as shown in the Table above. Conditions F and G produced the best results.
Conclusion
Direct PCR with the KAPA3G Plant PCR Kit is a quick and easy method for some plant species, however even with precise control over sample size, some sample types cannot be amplified successfully. On the other hand, crude sample PCR with the KAPA3G Plant PCR Kit is highly robust—even with the most challenging samples. Higher success rates can be achieved with crude PCR across a wide range of plant species and sample types, also enabling multiple PCRs from the same PCR and eliminating the need for precise control of sample size.
Generally, minimal optimization is required, provided that reaction conditions were optimized with purified DNA before attempting crude sample PCR. For those sample types that do require additional optimization, combinations of increased enzyme and MgCl2, as well as the use of 1 mM TCEP, can be evaluated to determine optimal reaction conditions. In exceptional cases, further optimization of cycling parameters may be necessary.
References
To continue reading please sign in or create an account.
Don't Have An Account?