PURedit™ Cas9 – A High Specificity and High Efficiency Genome Editing RNP System
- Why is SpCas9 Widely Used?
- Development of the CRISPR Gene Editing PURedit™ Cas9 RNP System
- Engineering Methods for Our Cas9 Protein Variants
- Identification of a Triple Mutant Cas9 Protein Yields Optimized Performance Characteristics
- PURedit™ CRISPR Cas9 RNP System and Related Reagents
Why is SpCas9 Widely Used?
While flexible and affordable, Streptococcus pyogenes CRISPR-Cas9 (SpCas9) is often used as a primary genome editing tool for research in diverse organisms, including plants and humans. Native SpCas9 tolerates the mismatch between the guide RNA sequence and target DNA sequence; however, this limitation often results in off-target effects. To alleviate this drawback, several groups have developed various SpCas9 variants with improved specificity through structure-based rational design or directed evolution. However, as these specificity-enhanced variants were selected under plasmid overexpression conditions, they often suffer from a substantial reduction in editing efficiency when they are delivered as a purified recombinant protein in combination with an in vitro transcribed or a chemically synthesized guide RNA in pre-assembled ribonucleoprotein (RNP) complexes. Here, we describe the development and performance evaluation of our PURedit™ RNP system, comprising of our high fidelity Cas9 Plus Protein and our PEXBUFF transfection enhancer.
Development of the CRISPR Gene Editing PURedit™ Cas9 RNP System
In order to maintain specificity and activity, we explored a different protein engineering strategy. This strategy included identifying and mutating critical residues that might be involved in different target DNA interactions or Cas9 nuclease activation modalities and evaluating their performance. We screened these variants in human cells using purified recombinant protein in complex with synthetic single guide RNA (sgRNA). By stratifying protein variants on a set of target and off-target sites, we identified several protein variants that possess balanced specificity, and favorable, single mutations. These variants were then trialed in combinations to identify the mutations that optimized both on-and off-target activities. In a parallel effort, we screened various compounds for their ability to enhance Cas9 RNP complex editing efficiency and identified an RNP enhancer that can increase the editing efficiency of these high specificity variants on difficult-to-edit genomic sites. Thus, by combining one of the high specificity variants with the RNP enhancer, we have developed a PURedit™ Cas9 RNP system for genome editing applications when Cas9 RNP complex delivery is desired.

Figure 1. Purified Cas9 protein evaluation approach for both on-and off-target activities. Cas9 variants harboring targeted mutations were expressed in E. coli BL21AI by autoinduction and purified by Ni-NTA affinity chromatography. Purified Cas9 proteins were evaluated with synthetic sgRNAs by RNP complex delivery for their on-target and off-target activities in human K562 cells. Multiple protein engineering cycles were performed to identify variants with balanced activity and specificity.
Engineering Methods for Our Cas9 Protein Variants
We employed a multimodality strategy to introduce mutations on SpCas9 protein residues that might be involved in electrostatic interactions with the sugar-phosphate backbone of the non-target strand DNA, guide RNA/target strand DNA heteroduplex stability, and REC3 domain-mediated HNH nuclease activation. Furthermore, in contrast to previous Cas9 specificity improvement efforts that invariably mutated the residues to alanine, regardless of their structural or functional characteristics, we employed a structurally similar substitution strategy to minimize the reduction in activity. The overall protein engineering workflow is presented in Figure 1. In parallel, we screened an array of compounds to identify Cas9 RNP complex electroporation enhancers.

Figure 2. Activity and specificity evaluation of engineered Cas9 recombinant protein variants.The activity of a set of engineered Cas9 protein variants on a human β-globin gene (HBB) target and a single mismatch off-target in K562 cells. Several variants exhibited high activity and high specificity while some lost nearly all the activity.
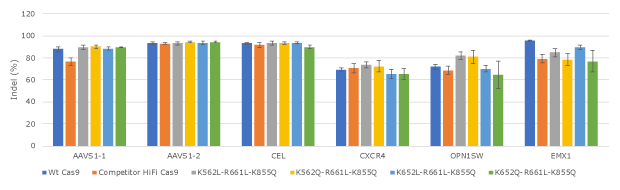
Figure 3. Evaluation of lead Cas9 protein variants for editing efficiency at expanded set of loci in human K562.All Cas9 proteins were tested in three replicates in human K562 cells by electroporation.
Identification of a Triple Mutant Cas9 Protein Yields Optimized Performance Characteristics
Improving Cas9 specificity while maintaining its activity is a delicate balancing act. Our data suggests that mutating a relevant residue to other structurally similar residues is more likely to achieve this balance. We also found that some single and double mutants could significantly improve the specificity and maintain the wild-type activity; however, they can still incur high levels of off-target effects on single-mismatch off-target sites. Therefore, we explored triple and quadruple mutations in our later engineering cycles and identified a set of triple mutants that can effectively minimize single mismatch off-target effects while maintaining a comparable wild-type activity level (Figures 2-4). Combining one of the triple mutants with a Cas9 RNP enhancer, we developed a PURedit™ Cas9 RNP system.
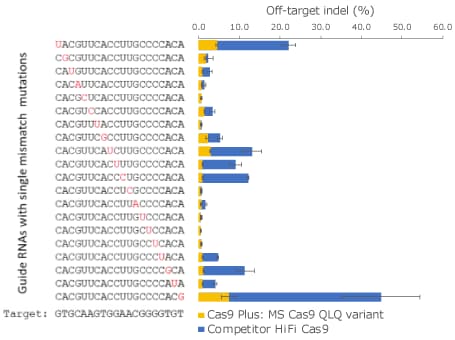
Figure 4. Specificity comparison between the K652Q-R661L-K855Q variant and a competitor HiFi Cas9.Both Cas9 proteins were tested in three replicates in human K562 cells with 20 synthetic sgRNAs, each harboring a single mismatch on a different position against the same human HBB target site as presented in Figure 2.
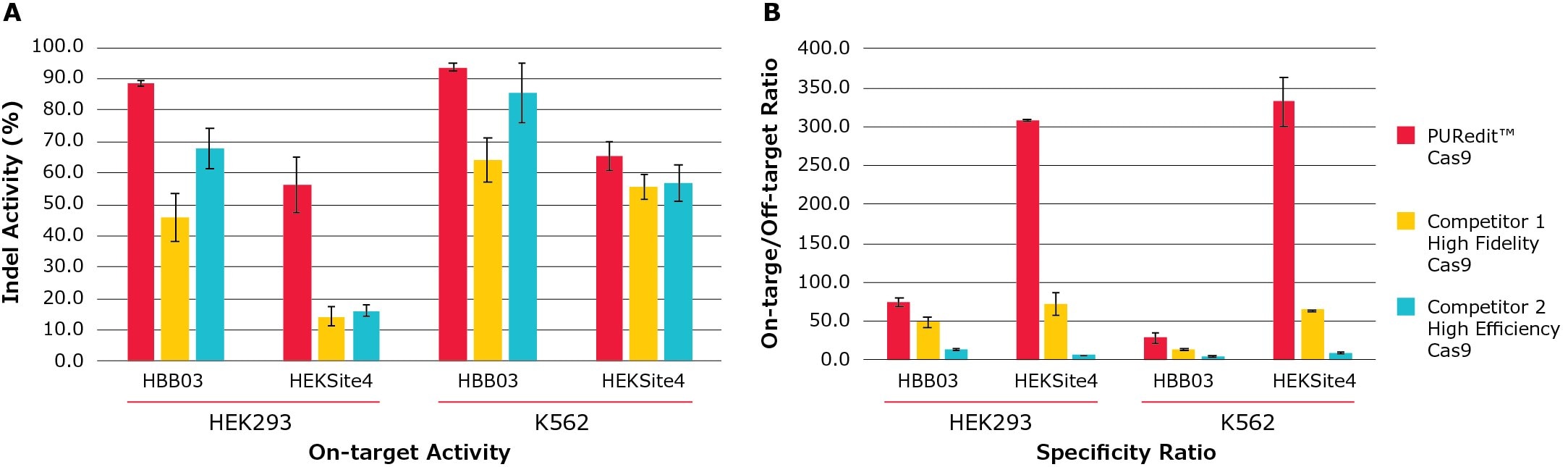
Figure 5. PURedit™ CRISPR RNP on-target activity and specificity ratio compared to competitors.PURedit™ Cas9, competitor 1 high fidelity variant Cas9, and competitor 2 high efficiency Cas9 were each complexed with PURedit™ sgRNAs and electroporated into both HEK293 and K562 cells. Two PURedit™ sgRNAs targeting two different genes were used with each Cas9 variant. Each transfection contained 5 µg of Cas9 protein and 100 pmol of PURedit™ sgRNA. (A) The average percentage of insertions and deletions for the two sgRNAs at their target sites (Indel Activity %) was determined by next generation sequencing (NGS). At both target sites in both cell lines, the PURedit™ RNP complexes showed greater cutting efficiency than competitors. (B) Transfected cells from (A) were analyzed for specificity by quantifying the insertions and deletions at the most active off-target sites, and the ratio of on-target to off-target activity was calculated. PURedit™ Cas9 shows the highest ratio at all targets, confirming high specificity and activity for the intended target sites.
PURedit™ CRISPR Cas9 RNP System and Related Reagents
We developed a set of high specificity SpCas9 protein variants that maintain high levels of activity through multimodal protein engineering and RNP complex delivery. We also identified a Cas9 RNP electroporation enhancer to increase Cas9 RNP editing efficiency, particularly on difficult-to-edit genomic sites. By combing one of the high specificity variants with the RNP enhancer, we developed a PUReditTM RNP system to enable efficient genome editing with greatly reduced off-target effects as compared with other commercial high fidelity or high efficiency Cas9 protein products (Figure 5).
For additional resources, explore our predesigned CRISPR gRNAs, design a custom gRNA or select a custom gRNA that best matches your needs. For all additional inquiries, please contact one of our experts.
Related Materials
To continue reading please sign in or create an account.
Don't Have An Account?