Reliable DNA extraction from Whatman® FTA® cards
This study examined the yield and quality of DNA from samples applied to Whatman® FTA® cards, using five common methods of DNA extraction. The data demonstrate that all five methods yield purified DNA of sufficient quantity and quality for both quantitative PCR and short tandem repeat analyses. In addition, EasiCollect™ device for buccal sample collection was shown to be compatible with all five DNA extraction methods, and provided final DNA yields equal to or higher than those from separate foam swabs.
Introduction
Whatman® FTA® family of products (Figure 1) facilitates collection, transport, purification, and long-term, room-temperature storage of nucleic acids, all in a single device. FTA technology has the ability to lyse cells on contact, denature proteins, remove contaminants, and protect DNA from degradation. In the classic scenario for FTA cards, purified DNA is analyzed while still bound to the matrix (i.e., using the “punch-in” method). This study examined an alternative option for FTA cards—DNA extraction into solution.
Materials and Methods
Sample collection
Whole blood and buccal cells were collected onto FTA cards.
Blood
Blood from ten unrelated individuals was collected by venipuncture into vacuum collection tubes containing dipotassium EDTA. Blood (40 μL) was applied to each FTA Micro Card. Cards were dried for at least 3 h at room temperature.
Figure 1. Whatman® FTA® family of products.
Buccal samples
Buccal samples were collected from ten unrelated individuals, using either EasiCollect devices from Cytiva (Figure 2) or foam swabs. Samples were applied to FTA cards according to the manufacturer’s instructions. Cards were dried for at least 2 h at room temperature.

Figure 2. EasiCollect™ is an all-in-one device that collects buccal samples and transfers them to an integrated FTA card.
Methods for DNA extraction
DNA was extracted or isolated from FTA cards using five different methods, exactly as described in the following protocols. These methods include:
- Organic extraction
- Chelex 100 resin (142-1253, Bio-Rad Laboratories,vHercules, CA, USA)
- QIAamp™ DNA Investigator Kit (56504, Qiagen, Valencia, CA, USA)
- illustra™ tissue and cells genomicPrep Mini Spin Kit (Cytiva)
- DNA IQ™ Kit (TB297, Promega Corp, Madison, WI, USA)
Organic extraction of DNA
Prepare extraction buffer and Proteinase K stock solution. Extraction buffer: 10 mM Tris-HCl, pH 8.0; 10 mM EDTA, disodium salt, pH 8.0; 100 mM sodium chloride; and 2% v/v SDS. Proteinase K stock solution: 20 mg/mL in sterile distilled water.
- Use a Harris Uni-Core disposable punch (1.2, 2.0, 3.0, or 6.0 mm) to remove a disc from the center of a dried sample spot. Place the disc in a clean RNase/DNase-free 1.5 mL microcentrifuge tube.
- Add 500 μL of extraction buffer and 20 μL of Proteinase K stock solution.
- Vortex and incubate at 56 °C overnight with agitation.
- Add an equal volume of buffered phenol (pH 8.0), vortex briefly, and centrifuge for 10 min at maximum speed in a microcentrifuge.
- Transfer the upper, aqueous phase to a new microcentrifuge tube that contains 500 μL of chloroform.
- Vortex thoroughly and centrifuge for 10 min at maximum speed in a microcentrifuge.
- Transfer the upper, aqueous phase to a new tube containing 50 μL of 3 M sodium acetate, pH 5.2.
- Add 800 μL of 100% ethanol, vortex, and precipitate at -20 °C for at least 1.5 h.
- Recover DNA by centrifuging for 30 min at maximum speed in a microcentrifuge and decanting the supernatant.
- Add 1 mL of 70% ethanol to the pellet and centrifuge for 20 min at maximum speed in a microcentrifuge.
- Dry the pellet in a vacuum centrifuge for 30 min.
- Dissolve the pellet in 50 μL of TE buffer or water.
DNA isolation using Chelex 100 resin
Prepare a 5% w/v suspension of Chelex 100 resin in sterile water prior to use; stir to maintain a suspension.
- Use a Harris Uni-Core disposable punch (1.2, 2.0, 3.0, or 6.0 mm) to remove a sample disc from the center of a dried sample spot. Place the disc in a clean RNase/DNasefree 1.5 mL microcentrifuge tube.
- Macerate the FTA disc with a 20-gauge needle.
- Wash the disc by adding 1 mL of sterile water to each sample tube and incubating at room temperature for 10 min with occasional vortexing. Remove the water.
- Repeat the wash by adding 1 mL of sterile water to each sample tube and incubating at room temperature for 10 min with occasional vortexing.
- Centrifuge samples for 3 min at 20 000 x g.
- Remove and discard the supernatant.
- Add 200 μL of Chelex 100 suspension using a large-bore pipette tip.
- Incubate the sample at 56 °C for 20 min.
- Vortex for approximately 15 sec.
- Incubate at 100 °C for 8 min.
- Vortex for approximately 15 sec.
- Centrifuge the sample for 3 min at 20 000 x g.
- Transfer the supernatant to a new sterile pre-labeled microcentrifuge tube, being careful to avoid transfer of the Chelex 100 resin pellet.
- Store DNA at 2 °C –8 °C for short periods or at −20 °C for extended periods.
DNA isolation using QIAamp DNA Investigator Kit
Reagents and columns are provided with the kit.
- Use a Harris Uni-Core disposable punch (1.2, 2.0, 3.0, or 6.0 mm) to remove a sample disc from the center of a dried sample spot. Place the disc in a clean RNase/DNase-free 1.5 mL microcentrifuge tube.
- Add 280 μL of ATL buffer to the tube.
- Add 20 μL of Proteinase K and vortex for 30 sec.
- Heat for 60 min at 56 °C, vortexing for 30 sec every 10 min.
- Centrifuge at maximum speed in a microcentrifuge for 30 sec.
- Add 300 μL of AL buffer and vortex for 10 sec.
- Incubate at 70 °C for 10 min, vortexing for 10 sec every 3 min.
- Centrifuge at maximum speed for 30 sec and transfer the lysate to a minElute column.
- Centrifuge at 6000 x g for 1 min and discard the flowthrough.
- Add 700 μL of AW2 buffer and centrifuge for 1 min at 6000 x g. Discard the flowthrough.
- Add 700 μL of 100% ethanol and centrifuge for 1 minute at 6000 x g. Discard the flowthrough.
- Centrifuge at 20 000 x g for 3 min, then open the column lid for 10 min at room temperature.
- Add 50 μL of distilled water and incubate for 5 min at room temperature.
- Centrifuge at 20 000 x g for 1 min. Store the flowthrough, which contains the extracted DNA, at 4 °C until needed.
DNA isolation using illustra tissue and cells genomicPrep Mini Spin Kit
Reagents and columns are provided with the kit. Phosphate-buffered saline (PBS) is also required.
- Use a Harris Uni-Core disposable punch (1.2, 2.0, 3.0, or 6.0 mm) to remove a sample disc from the center of a dried sample spot. Place the disc in a clean RNase/DNasefree 1.5 mL microcentrifuge tube.
- Add 1 mL of PBS and centrifuge at 16 000 x g.
- Macerate the disc using a 20-gauge syringe needle.
- Centrifuge for 10 sec at 2000 x g.
- Add 50 μL of buffer 1 to each sample.
- Add 10 μL of Proteinase K to each sample and vortex for 15 sec.
- Incubate for 1 h at 56 °C
- Centrifuge for 10 sec at 2000 x g. Add 5 μL of RNase A (20 mg/mL). Incubate for 15 min at room temperature.
- Add 500 μL of buffer 4 and vortex for 15 sec.
- Incubate for 10 min at room temperature.
- Pipette the sample into a mini column (placed in a collection tube) and centrifuge for 1 min at 11 000 x g. Discard the flowthrough.
- Add 500 μL of buffer 4 to each column. Centrifuge for 1 min at 11 000 x g and discard the flowthrough.
- Place the column into a collection tube and add 500 μL of buffer 6 to the column.
- Centrifuge for 3 min at 11 000 x g. Transfer the column to a new 1.5 mL tube.
- Add 50 μL of pre-warmed elution buffer (buffer 6) to the column.
- Incubate for 1 min at room temperature.
- Centrifuge for 1 min at 11 000 x g. Discard the column and store the DNA at 4 °C until needed.
DNA Isolation using DNA IQ Kit
Reagents and columns are provided with the kit. A magnetic separation stand is also required.
- Use a Harris Uni-Core disposable punch (1.2, 2.0, 3.0, or 6.0 mm) to remove a sample disc from the center of a dried sample spot. Place the disc in a clean RNase/DNase-free 1.5 mL microcentrifuge tube.
- Add 100 μL of lysis buffer and incubate at 70 °C for 30 min.
- Transfer the contents to a spin column seated in a 1.5 mL tube and centrifuge at maximum speed for 2 min. Discard the spin column.
- Vortex the stock resin until thoroughly mixed, and add 7 μL of resin for the 1.2 mm disc, 7 μL for the 2.0 mm disc, 16 μL for the 3.0 mm disc, or 28 μL for the 6.0 mm disc.
- Vortex tubes for 3 sec and incubate at room temperature for 5 min, vortexing for 3 sec once every min.
- Vortex the tube for 2 sec and place on a magnetic stand.
- Decant the supernatant without disturbing the pellet.
- Add 100 μL of lysis buffer. Remove the tube from the magnetic stand and vortex for 2 sec.
- Return the tube to the magnetic stand and decant the lysis buffer.
- Add 100 μL of 1× wash buffer. Remove the tube from the stand and vortex for 2 sec.
- Return the tube to the magnetic stand and decant the wash buffer.
- Repeat wash steps 10 and 11 two more times for a total of three washes. Decant the wash buffer following the final wash.
- With the tube in the magnetic stand, open the lid and air-dry for 10 min.
- Add 100 μL of elution buffer. Close the lid and vortex for 2 min. Incubate for 5 min at 65 °C.
- Remove the tube from the heat and vortex for 2 sec, then immediately place the tube in a magnetic stand.
- Transfer the eluate to a fresh tube and store the eluted DNA at 4 °C until needed.
DNA analyses
DNA extracted from FTA cards was measured using quantitative PCR (qPCR). DNA was subjected to short tandem repeat (STR) analysis to assess its quality.
DNA quantitation by qPCR
Extracted DNA was measured using quantitative PCR (qPCR) and an ABI™ 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Reactions were prepared using the Quantifiler™ Human DNA Quantification Kit (4343895, Applied Biosystems). The DNA quantitation assay combines two 5’ nuclease assays. The first is a target-specific human DNA assay consisting of two primers for amplifying human DNA and one TaqMan™ MGB probe labeled with FAM™ dye for detecting the amplified sequence. The second assay is an internal PCR control (IPC) assay consisting of two primers for amplifying the IPC template DNA and one TaqMan MGB probe labeled with VIC™ dye for detecting the amplified IPC DNA. Standard curves for DNA quantitation were prepared using control DNA supplied with the kit.
Short tandem repeat analysis
STR analysis of extracted DNA was conducted using the Powerplex™ 16 System (Promega) according to the manufacturer’s instructions. Amplification was performed in a 9700 thermal cycler (Applied Biosystems), and amplification products were analyzed on an ABI PRISM™ 3130XL Genetic Analyzer (Applied Biosystems) according to the manufacturer’s instructions.
Results and discussion
DNA quantitation by qPCR
DNA was extracted from FTA discs containing blood or buccal cells from ten unrelated individuals. Four sizes of disc were used for each extraction method, and the total quantity of extracted DNA per disc was determined as described in Materials and Methods.
For each of the five extraction methods, a minimum of 10 ng of genomic DNA was recovered from blood samples on FTA cards (Figure 3). The amount of DNA extracted using each method increased as the disc size increased.
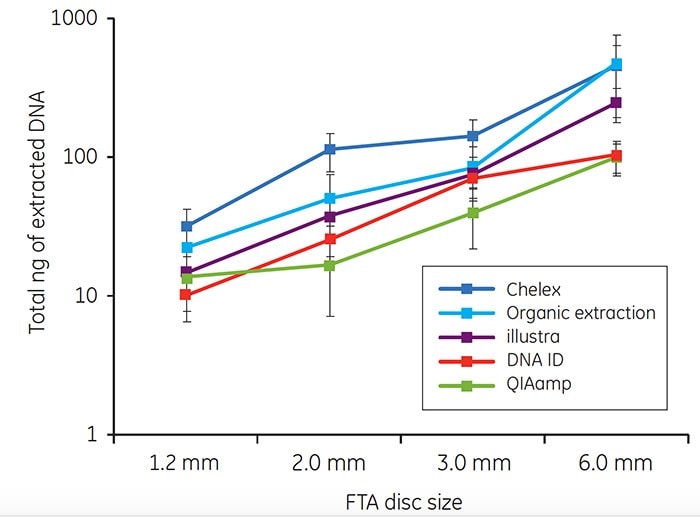
Figure 3. Total DNA (ng) isolated from discs taken from the center of FTA cards spotted with 40 μL of whole human blood. Each point on the graph represents the mean recovery of DNA from a single disc from ten individuals. Error bars represent standard deviations. Note that the y axis represents a logarithmic scale.
Buccal samples have historically produced greater variability in DNA yield than blood samples. The variability in buccal DNA yield is most likely due to clumping of cells on the matrix. As for blood samples, the amount of DNA recovered with each extraction method tended to increase as the disc size increased. Note that higher DNA yields were often obtained from buccal samples collected with EasiCollect devices than from samples collected with foam swabs (Figure 4). The data shown in Figures 3 and 4 illustrate that commonlyused DNA extraction methods and kits do provide DNA of sufficient quantity and quality for successful qPCR.
DNA extracted from collected buccal samples
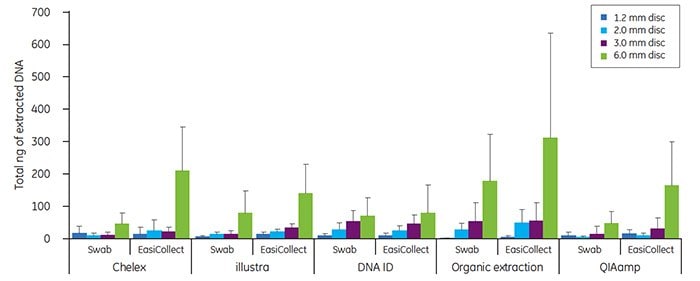
Figure 4. Total DNA (ng) isolated from discs taken from the center of an FTA card containing buccal cells, applied with either a foam applicator or an EasiCollect device as indicated,
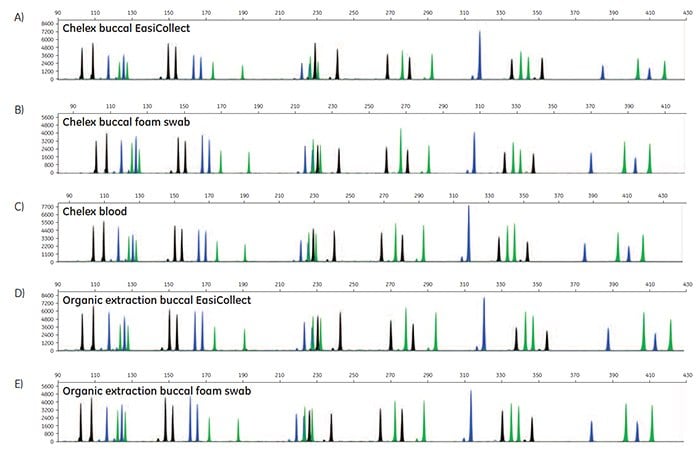
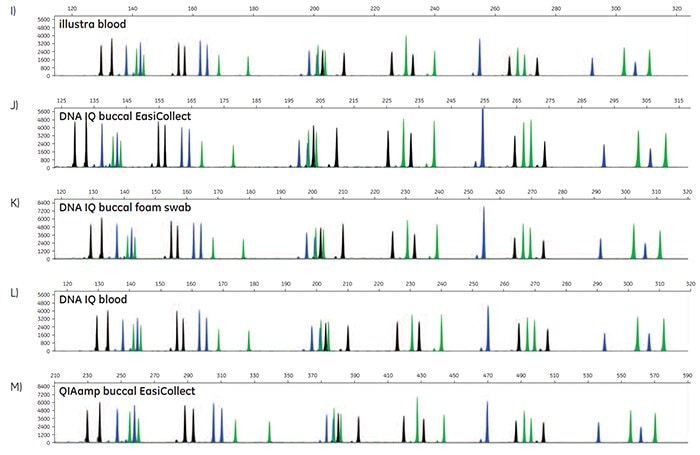
Figure 5. A-O. STR profiles of DNA extracted from FTA cards containing blood or buccal samples from Test Subject 1.
STR data were generated to determine the accuracy of allele calls for DNA extracted from FTA cards. For each DNA extraction method, duplicates of DNA from FTA cards were run for three blood samples, three buccal samples collected using EasiCollect devices, and three buccal samples collected using separate foam swabs, resulting in a total of 18 samples per DNA extraction method. Note that the three samples were from three test subjects. Of the 90 STR profiles collected, 87 samples provided quality data with accurate allele calls on the first attempt; three samples required a reinjection for 100% accurate allele calls. These data illustrate that commonly-used DNA extraction methods and kits provide DNA of sufficient quantity and quality to support allele call accuracy as high as 100% in STR analysis.
Conclusions
This study shows that five common DNA extraction methods yielded enough DNA from FTA cards for downstream quantitative analysis. This DNA was of sufficient quality for determining human identity via STR profiling. In addition, collection of buccal cells using the EasiCollect device often resulted in a higher yield of DNA than collection using a foam swab. These data demonstrate that the EasiCollect device is compatible with current validated laboratory protocols for DNA extraction.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?