Sanger Whole Genome CRISPR Libraries
Genome-wide loss-of-function screening is a powerful approach to discover genes and pathways that underlie biological processes. To help you unravel your genes and pathways of interest, we collaborated with The Wellcome Sanger Institute to make the first arrayed lentiviral CRISPR knockout libraries for human and mouse genomes.1 Covering over 17,000 human and over 20,000 mouse genes with two optimized gRNAs for each of them, our Sanger Whole Genome Arrayed Libraries are powerful tools that can help you make the next exciting discovery.
Arrayed Screens are Effective and Versatile, even in Difficult-to-transfect Cell Lines
While pooled CRISPR libraries can provide genome-wide knockout, an arrayed approach offers many benefits:
- Enriched phenotypic content: flexibility in functional assays and analysis possibilities beyond viability—use cell imaging, fluorescence, luminescence, and colorimetric readouts
- Streamlined data analysis: easy hit identification with no requirement for deconvolution
- High sensitivity: a single target in every well provides better signal-to-noise ratio
- Ease of identifying candidate genes: closer genotype to phenotype correlation
- Flexibility: can be applied to a wider range of cell types including primary cells and neurons
Our Sanger Arrayed CRISPR Libraries are exclusive to the Sigma-Aldrich® portfolio, and are available off-the-shelf. They can be combined with other tools to get the results you need: perform initial pilot experiments and optimize your cellular system and assays with our CRISPR controls, or use the arrayed libraries as part of complementary experiments with our existing CRISPR, RNAi and ORF libraries for validation or rescue.
Start Your CRISPR Screen Today
Why troubleshoot false positive results when you can use the gRNA-optimized, arrayed Sanger CRISPR libraries? Our exclusive Sanger CRISPR library provides extensive, high-quality gene knockout, so you can fast-track your research with the first whole genome arrayed library.

Our lentiviral-based CRISPR screens offer many options in their application:
- Stable or transient transfection
- Enrich population of cells (vectors allow puromycin- and BFP-based selection)
- BFP tag allows visualization of positive cells (Figure 1)
- Create a stable cell line with viral particles for sustained integration
- Transduce virtually any cell type (both dividing and non-dividing cell lines)
- Get custom titers, suitable for any application
Figure 1. BFP expression in A549-Cas9-Blast stable cells infected with arrayed CRISPR lenti-gRNA viruses, seven days post infection, 20X magnification.
Need help with CRISPR fundamentals? Learn more about Getting Started with CRISPR.

Sanger Library Specs
Glycerol Format
- Approximately 400 96-well plates, 50 µL bacterial glycerol stock per clone
- Human or mouse species
Lentiviral Format
- Approximately 100 384-well barcoded labeled Costar plates with pierceable aluminum seals
- 10 uL per clone at a minimum titer of 1x106 particles/mL as determined by p24 assay
- Human or mouse species
Sanger CRISPR Guide RNA Design and Vector Features
gRNA Design
- Maximizes gene knockout by targeting the first half of the protein coding sequence
- Optimizes targeting of all transcripts
- Avoids the first 90 bp of coding sequence where alternative start codons often reside
- Targeted to highly conserved sequences avoiding SNPs, to ensure representation in multiple cell lines
- Stringent rules reduce or eliminate the potential for off-target effects
Vector Features
- Simplify the workflow with puromycin selection
- Visualize CRISPR-expressing cells and enrich the transduced population with BFP
- Transposase sites provide an alternative to lentiviral delivery
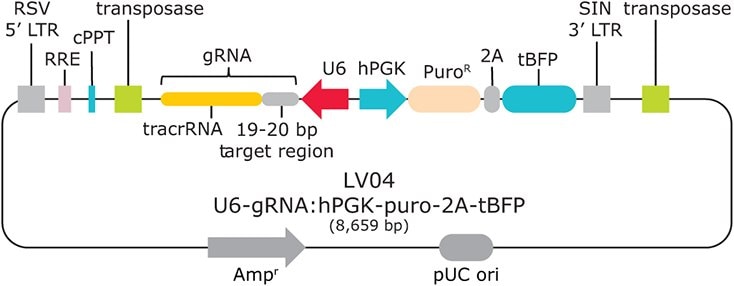
Figure 2.Sanger Lentiviral CRISPR vector schematic (LV04)
Lentiviral Production Expertise
We are committed to providing unmatched manufacturing and support for genome editing technology. With state-of-the-art robotics, liquid handling, LIMS and dedicated laboratories we provide superior quality and convenient service for your CRISPR research needs.
High quality manufacturing systems and personnel:
- High throughput (HT) robotic handling
- 1D and 2D barcoding
- Highly trained experts with 10> years of lentiviral gene editing experience
- High-throughput viral production
From single targets to whole genomes
The Sanger library is the only tool that allows both whole genome CRISPR KO interrogation and sophisticated phenotypic readout in a convenient arrayed format. For smaller- scale experiments, benefit from our manufacturing quality and the design expertise of the Wellcome Sanger Institute to build a custom panel for your research.
Do you need more focused or affordable screens?
- Choose a single Sanger QuickPick™ clone for any gene
- Cherry-pick a custom Sanger panel of any size
- Order any gRNA design in the Sanger vector
- Target any pathway or gene family with a Sanger panel
|
|
Don’t see the option you need? Specify your own gRNA for manufacturing in the Sanger backbone. From whole genome libraries to individual clones.
Sanger screen gRNAs are optimized and facilitate robust gene editing. Designed to maximize knockouts, the gRNAs are highly effective as assessed by CEL-I assays for indel detection. In addition, two gRNAs are provided for each gene, allowing candidate stratification following a whole-genome screen, maximizing efficiency and reducing false positives.
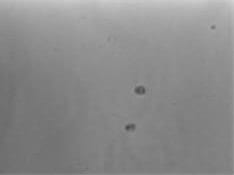
A.
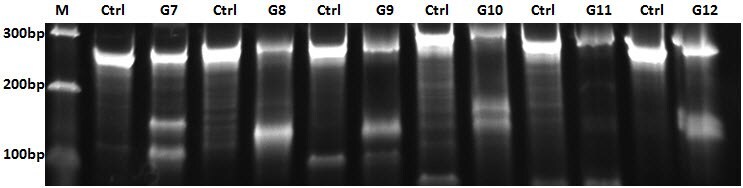
B.
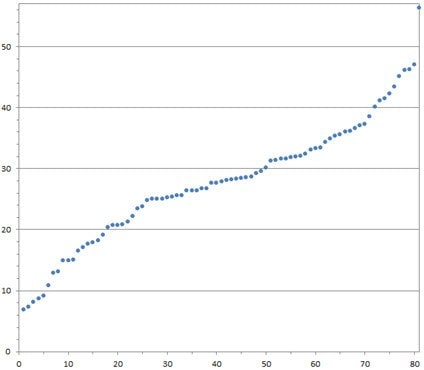
C.
Figure 3. Clones from the Sanger lenti-gRNA library demonstrate definitive gene editing in a CEL-I assay for mutation detection. A549-Cas9-Blast cells were infected with Sanger screen gRNAs, selected with puromycin, then subjected to a CEL-I assay. A) Cell growth after puromycin selection for four days; B) Representative agarose gel from CEL-I assay showing DNA cleavage in samples from cells that received Sanger guides (lanes G7-G12) vs. no cleavage in controls; C) A plot of % DNA cleavage in the CEL-I assay (y-axis) for a representative selection of gRNA clones (each dot represents a single clone) showing high average nuclease activity and high active gRNA frequency.
See how the designer of the Sanger screen is using this tool to help find new therapies for neurodegenerative diseases.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?