Restriction Endonucleases - The Molecular Scissors
Background
The term “Restriction enzyme” originated from the studies of Enterobacteria phage λ (lambda phage) in the laboratories of Werner Arber and Matthew Meselson. The ability of certain E. coli strains to inhibit the activity of lambda phage by the enzymatic cleavage of the phage DNA was studied and the enzyme responsible for this growth restriction was termed a restriction enzyme.1, 2, 3
Werner Arber, Daniel Nathans, and Hamilton O. Smith were awarded the Nobel Prize for Physiology or Medicine in 1978 for their discovery and characterization of restriction enzymes, which led to the development of recombinant DNA technology.
Introduction
Restriction enzymes are also called "molecular scissors" as they cleave DNA at or near specific recognition sequences known as restriction sites. These enzymes make one incision on each of the two strands of DNA and are also called restriction endonucleases.4
Viruses infect the host cells by injecting their DNA into the cells. This viral DNA hijacks the host cell’s machinery for reproduction of viral progeny, resulting in the host cell’s death. To overcome the viral infection, many bacteria and archaea have evolved several mechanisms. A major protective mechanism involves the use of restriction enzymes to degrade the invading viral DNA by cleaving it at specific restriction sites. At the same time, the host cell protects its own DNA from being cleaved by employing other enzymes called methylases, which methylate adenine or cytosine bases within host recognition sequences. For each of the restriction enzyme, the host cell produces a corresponding methylase that methylates and protects the host DNA from degradation. These enzymes make up the restriction-modification (R-M) systems.
The restriction enzymes catalyze the hydrolysis of the bond between the 3’-oxygen atom and the phosphorus atom in the phosphodiester backbone of DNA. The enzymes require Mg2+ or other divalent ions for their activity.
Nomenclature
Smith and Nathans suggested the naming guidelines for restriction endonucleases in 1973. According to these guidelines, the names of the enzymes begin with an italicized three-letter acronym. The first letter indicates the first letter of the bacterial genus from which the enzyme has been isolated and the next two letters are derived from the bacterial species. These may be followed by extra letters or numbers to indicate the serotype or strain. This is followed by a space and a Roman numeral to indicate the chronology of identification. For example, Hind III was the third of four enzymes isolated from Haemophilus influenza serotype d.6
Types of Restriction Enzymes
Based on the composition, characteristics of the cleavage site, and the cofactor requirements, the restriction endonucleases are classified into four groups, Type I, II, III, and IV.
The site of cleavage is indicated by the red arrow in the table.
Factors Affecting the Activity of Restriction Enzymes
Depending on the substrate DNA and the reaction conditions, restriction enzymes show a wide variation of cleavage and possible star activity. In order to obtain the desired cleavage, it becomes important to control the following factors:
- Star activity: Under sub-optimal reaction conditions, some restriction enzymes cleave base sequences at sites different from the defined recognition sequence. In other words, they cleave at non-specific sites. This phenomenon is called star activity. Some of the factors that induce star activity are high salt and glycerol concentration, presence of impurities, excessive enzyme compared to substrate DNA, increased incubation time, or incompatible buffer and cofactor.
- Methylated DNA: Several DNA molecules are methylated at the recognition site, making them resistant to cleavage by certain restriction enzymes. For example, most E. coli strains express Dam or Dcm methyltransferases that methylate specific recognition sites to form G6mATC and C5mCA/TGG, respectively. G6mATC is resistant to cleavage by Mbo I.
- Temperature: Most endonucleases optimally digest the target DNA at 37 °C. However, there are some exceptions with lower or higher optimal temperatures. For example, Taq I optimally digests at 65 °C and Apa I (Catalog No. 10899208001) digests at 25 °C.
Isoschizomers and Neoschizomers6
Isoschizomers are restriction enzymes with the same recognition sequence and cleavage sites. Example: Sph I (CGTAC/G) and Bbu I (CGTAC/G)

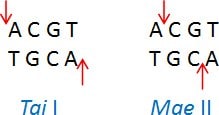
Neoschizomers are restriction enzymes with the same recognition sequence but cleave the DNA at a different site within that sequence. Example: Tai I (ACGT/) and Mae II (A/CGT)
Products of Restriction Digestion
Restriction digestion of double-stranded DNA produces two kinds of ends: Sticky ends and Blunt ends.
Blunt ends possess a 5’-phosphate group that promotes ligation. They are universally compatible with other blunt-ended DNA.
Blunt ends generated by EcoR V

Sticky ends are small stretches of single-stranded DNA capable of self-ligation or ligation with a complementary region from another DNA molecule. The sticky ends possess 3’- or 5’-overhangs of 1–4 nucleotides.
5’ Cohesive end generated by Bln I (Catalog No. 11558170001)

3’ Cohesive end generated by Kpn I

Buffer System
Our restriction enzyme collection has been optimized for digestion using five unique buffers. When digesting DNA using a single enzyme, use the buffer supplied with the enzyme. For double digestion of DNA, use the buffer in which both the enzymes show 100% activity. Alternatively, the optimal buffer can be determined from the chart of common double digestions. In some cases, sequential digestion is recommended due to buffer incompatibility (composition or temperature). For protocols on restriction digestion with single enzyme, two restriction enzymes and sequential DNA Digestion, refer to Restriction Enzyme Digest Protocol. The selection of the correct buffer is crucial for obtaining high activity of both enzymes and for avoiding star activity. The choice of five buffers allows the user to select compatible buffers for the desired digestion. Most digestions require 1–2 buffers, which becomes cost-effective.
Applications
The ability of restriction endonucleases to cleave DNA at specific recognition sites has enabled extensive use of these enzymes as essential tools in several molecular biology techniques. Some of the major applications are explained below:
- Molecular cloning: A popular application of restriction enzymes has been in the generation of recombinant DNA molecules. The process involves the cutting of the donor DNA (usually a plasmid) and the vector DNA (usually a gene from another organism) by a restriction enzyme to yield compatible ends. These ends could be either ‘blunt’ or ‘sticky’. The two cleaved DNAs are joined together using an enzyme called DNA ligase to generate a recombinant DNA molecule. This recombinant DNA can, then, be introduced into a host organism for replication. For more details, refer Restriction Enzyme Cloning Manual
- DNA mapping, also known as restriction mapping, involves the use of restriction endonucleases to obtain structural information of the DNA fragment or genome. Mapping involves determination of the order of the restriction enzyme sites in the genome. The DNA of interest, whose structure is to be determined, is cleaved with a series of restriction endonucleases to produce DNA fragments varying in size. These fragments are separated on an agarose gel to determine the structure of the DNA of interest.
Based on the known restriction enzyme sites of a specific DNA fragment, restriction endonucleases can be used to verify the identity of that DNA fragment. - Restriction landmark genomic scanning is a genome analysis method that utilizes a combination of restriction enzymes to visualize differences in methylation levels across the genome of a given organism. It is a useful technique to identify deviations from normal in any DNA. It is very effective in detecting hyper/hypomethylation in tumors, deletions or amplifications of genes, or changes in gene expression throughout the development of an organism.8
- Gene sequencing: A large DNA molecule can be sequenced by digesting it with restriction enzymes and processing the resulting fragments through a DNA sequencer.
- Restriction fragment length polymorphism (RFLP) involves the digestion of a DNA sample using restriction enzymes, separating these fragments based on length by gel electrophoresis and transferring them onto a membrane. These fragments are then bound to a radioactive or fluorescent labeled probe targeting specific sequences that are bracketed by restriction enzyme sites. An RFLP occurs when the resulting fragment lengths vary between individuals. Each individual has a unique pattern called the ‘biological bar code’. This technique was the first DNA profiling technique used in gene mapping, localization of genes for genetic disorders, determination of risk for disease, and paternity testing.9
- Pulse field gel electrophoresis involves the separation of large DNA fragments, mainly the fragments resulting from digesting a bacterial genome with a rare-cutting restriction enzyme. The unique pattern produced is used to distinguish different strains of bacteria. It may be useful to identify a particular strain as the cause of a widespread disease.10, 11
- Serial analysis of gene expression (SAGE) is a technique that involves the quantitative and simultaneous analysis of a large number of transcripts in the form of small tags. Restriction enzymes are used as an anchoring enzyme and a tagging enzyme in this technique.12
- Restriction enzyme-mediated integration (REMI) involves the use of restriction enzymes to produce compatible cohesive ends in the genome for the insertion of a mixture of plasmid DNA that has been linearized with a restriction enzyme. The plasmid DNA is transformed into the host cell along with a restriction enzyme that facilitates the integration of the DNA into cognate restriction sites in the chromosomes. This technique is useful for genetic screens and for the insertion of genetic and molecular markers at particular points in the genome to identify interesting genes based on their mutant phenotypes.13
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?