Next-Gen RNA Interference with MISSION® shRNA
Introduction
The technology of RNA interference emerged in its earliest form following a 1998 study in Caenorhabditis elegans1 and has since rapidly evolved to its current form as a revolutionary tool for studying gene function, biological pathways, and the physiology of disease. Earlier studies in petunias demonstrated a phenomenon (termed co-suppression) where over-expression of a homologous transgene resulted in silencing of both the transgene and the endogenous gene.2 The later studies in C. elegans provided breakthrough experiments that clearly established that double-stranded RNA (dsRNA) interfered with gene function. This discovery was called RNA interference or RNAi.
Work and refinement of the RNAi technology has grown. We now know the basic mechanism of the endogenous RNAi pathway, that the pathway is present in most eukaryotes,3 and how cellular machinery can be harnessed to silence gene expression. Key rules and parameters for use of small interfering RNAs (siRNAs) less than 30 base pairs (bp) were developed that allow use of these dsRNA in mammalian cells4 without triggering the cell’s antiviral response mechanism. Upon transfection, synthetic siRNAs by-pass cleavage by the RNase enzyme Dicer (Figure 1) and are taken up by the RNAi-induced silencing complex (RISC). RISC unwinds the double strand siRNA and the activated complex with the associated anti-sense siRNA strand targets the homologous mRNA transcript for cleavage and subsequent degradation. The reduction in transcript level results in lowered levels of the target protein, resulting in phenotypic changes. Gene silencing or knockdown can be assayed at the mRNA transcript level using methods such as qRT-PCR or at the protein level via Western blotting, ELISA, and more recently developed mass spectrometry-based protein quantitation methods such as AQUA™ technology.
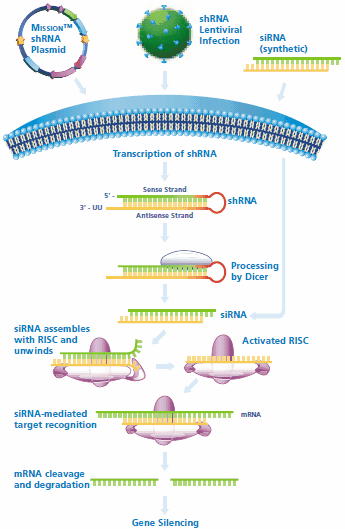
Figure 1.shRNA and siRNA mediated gene silencing. Cells may be directly transfected with MISSION shRNA plasmids for transient or stable gene silencing, while shRNA lentiviral particles may be used to transduce the cell line of interest. Following transcription of the shRNA in the nucleus, the hairpin enters the RNAi pathway when it is cleaved by Dicer to generate siRNA. The siRNA is recognized by RISC, which mediates cleavage of the target mRNA for gene silencing. Synthetic siRNA may be directly transfected and enters the RNAi pathway when it assembles with RISC.
Further advances have shown that siRNAs can be expressed from DNA vectors within the host cell, providing methods for longer term silencing, inducible silencing, and a plasmid DNA format that can be replicated for unlimited supply. In addition, these vector-based RNAi platforms may be integrated with viral delivery systems allowing the researcher to perform gene knockdown in a myriad of cell lines. Recent studies of endogenous microRNAs (miRNAs) suggested that synthetic miRNA mimics could be used to induce the RNAi pathway rather than directly using the standard 21 bp siRNA sequence. These synthetic forms of miRNA, termed short hairpin RNAs (shRNAs), are expressed from pol II or pol III promoters. The hairpin structure is recognized and cleaved by Dicer to form siRNA that is subsequently taken up by RISC for silencing of the target gene.
Discussion
As the portfolio of RNAi methods continues to expand, options become available for even the most complex systems being studied. Until recently, synthetic siRNA was the RNAi vehicle most broadly applicable to a wide variety of systems and applications. With commercial suppliers designing and producing synthetic siRNAs, little manipulation is required for the consumer. This format is amenable to any scale of research being performed provided the system is easily transfected (e.g., standard transformed cell lines). However, obstacles for using synthetic siRNAs include being a non-renewable resource, the transient nature of silencing, and the difficulty faced in transfecting primary cells and non-dividing cell lines such as neurons, lymphocytes, and macrophages. In addition, in vivo knockdown studies are particularly cumbersome.
For those facing the above hurdles, DNA vector-based shRNA methods provide the necessary solutions. shRNA expression vectors may be propagated in Escherichia coli and, thus, provide an unlimited supply of DNA for transfection. In addition, such vectors provide selectable markers for stable shRNA expression and gene silencing. One of the most attractive features of plasmid-based systems is the coupling of the technology to viral delivery systems. Vectors containing appropriate viral packaging signals and regulatory elements may be used to package the shRNA sequence into infectious virions. When appropriately pseudotyped, these viral particles can transduce a broader spectrum of cell lines and overcome issues faced in standard transfection methods. Viral delivery systems have been extensively studied for gene therapy research and have, thus, undergone numerous modifications for safety and use. It is important, however, to consider features of the various viral systems. Adenovirus and a number of retroviruses such as lentivirus and murine stem cell virus (MSCV) are a few of the commonly used viral delivery systems. Adenovirus utilizes receptor-mediated infection and does not integrate into the genome for stable silencing experiments, while MSCV cannot integrate into non-dividing cell lines such as neurons, etc. The lentiviral system, pseudotyped with the VSV-G envelope protein, presents the most attractive system for viral packaging and delivery of shRNA constructs. This is due to its broad tropism and non-receptor mediated delivery, its ability to integrate into the genome for stable gene silencing, and the fact that it does not require a mitotic event for integration into the genome; thus, extending its use to both dividing and nondividing cell lines. The lentiviral system is also not known to elicit immune responses minimizing concerns of off-target effects and use in in vivo applications.
In an effort to help further the development and distribution of tools for RNAi research, Sigma-Aldrich announced its membership and sponsorship of The RNAi Consortium (TRC) in March 2005. TRC is a research collaboration between The Broad Institute and renowned scientists and their laboratories from Harvard Medical School, Massachusetts Institute of Technology, The Whitehead Institute, Dana Farber Cancer Institute, Massachusetts General Hospital, Washington University, and Columbia University. Four other leading life science research organizations in addition to Sigma-Aldrich are also sponsoring members. The mission of The Broad Institute and TRC is to create comprehensive tools for genomics medicine, make them broadly available to scientists worldwide, and to pioneer applications of these tools for the study of disease. For three years, the consortium’s goal is to create a comprehensive library of RNAi reagents (vector-based shRNA clones) designed to knockdown expression of human and mouse genes, thereby, enabling scientists to elucidate gene function in normal physiology and disease.
As a scientific collaborator and commercial partner, Sigma-Aldrich will assist the development, manufacturing, and global distribution of TRC’s human and mouse lentiviral vector-based shRNA libraries (MISSION™ shRNA). The collection is designed and developed by the Broad Institute of MIT and Harvard and is in the process of being expanded to 150,000 clones targeting 15,000 annotated human genes (MISSION TRC-Hs1.0) and 15,000 annotated mouse genes (MISSION TRC Mm1.0). Approximately 50,000 clones targeting 7,100 human and 2,900 mouse genes are currently available. The libraries include a broad range of gene families, functional classes, and druggable targets.
MISSION® shRNA constructs are designed using a proprietary algorithm that scores potential sequences for efficient knockdown of the endogenous gene based on nucleotide content, position within the target gene, and sequence specificity via BLAST searches to minimize off-target effects. Up to five shRNA sequences are individually cloned into pLKO.1-puro (Figure 2) for broad coverage of each target gene and varying degrees of knockdown (Figures 3A and 3B). The hairpin structure includes an intramolecular 20-21 bp stem and 6 base loop that is recognized and cleaved by Dicer upon expression via the U6 (pol III) promoter in the host cell. The resulting siRNA duplex then continues in the RNAi pathway by association with RISC. The puromycin resistance marker is present for stable selection in mammalian cells, while the ampicillin resistance marker provides for plasmid propagation in E. coli.
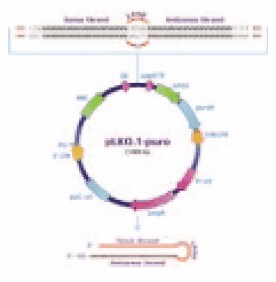
Figure 2.pLKO.1-puro vector.
The constructs may be used for transient or stable transfection of mammalian cells. In addition, pLKO.1-puro features allow for the generation of lentiviral particles for infection of a wide variety of cells. The 5′ long terminal repeat (LTR), SIN/LTR (3′ LTR), and Psi Packaging Signal permit viral packaging using 2-plasmid or 3-plasmid lentiviral packaging systems.5 Using this multiplasmid approach, resulting viral particles are replication incompetent and cannot be propagated, facilitating safe use of the particles.6,7 Also, a deletion in the U3 region of the 3′ LTR (SIN/LTR) does not affect generation of the viral genome during packaging, but results in loss of the transcriptional capacity of the viral LTR once transferred to target cells. This feature also helps reduce the risk of emergent replication competent viral particles and avoids problems linked to promoter interference. Most important, however, is the fact that the integrated U6 promoter and shRNA sequence may be stably expressed within the targeted cell line.
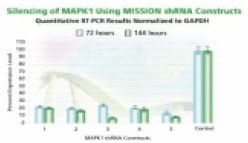
Figure 3A.MAPK 1 gene silencing. The Human Mitogen-Activated Protein Kinase 1 (MAPK 1, NM_138957) MISSION shRNA set (5 individual hairpins) was used to generate lentiviral particles in packaging cells. HEK 293T cells were infected with the lentiviral particles containing the shRNA sequences. 72 and 144 hours post-infection, total RNA was purified and analyzed with the appropriate TaqMan™ Gene Expression Assay. The difference in Ct between infected samples and uninfected control cells along with the efficiency of PCR were used to generate percentage expression levels. Percentage is expressed as a level of the control (100%).
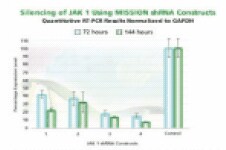
Figure 3B.JAK 1 gene silencing. The Human Janus Kinase 1 (JAK 1, NM_002227) MISSION shRNA set (4 individual hairpins) was used to generate lentiviral particles and infect HEK 293T cells as above. Total RNA from 72 and 144 hours post-infection was also analyzed using the same procedures. The results shown indicate the shRNA constructs produce JAK 1 gene silencing compared to control levels.
Summary
RNAi technology has been one of the key biological breakthroughs of the last decade and has revolutionized basic biology, gene function studies, and it also holds promise to revolutionize drug discovery and therapeutics. Synthetic siRNAs have delivered the genetic tools needed for studying eukaryotic systems that were previously difficult to study. However, in the quest for studying systems with varying degrees of complexity, obstacles were identified.
MISSION pre-cloned shRNAs overcome these issues by providing a system for long-term silencing and phenotypic observation, plasmid for unlimited propagation, options for transient or stable transfection, and the ability to generate lentiviral particles for infection and integration of primary cells, dividing cells, and non-dividing cells. Sigma-Aldrich is committed to providing unmatched infrastructure and service for your RNAi research needs.
The RNAi Consortium shRNA library is produced and distributed under license from the Massachusetts Institute of Technology.
References
To continue reading please sign in or create an account.
Don't Have An Account?