Small RNA and miRNA (microRNA) Sequencing
| Webinar - Decreasing miRNA Sequencing Bias Using a Single Adapter and Circularization Approach - Watch Now |
Next-Generation Sequencing of miRNA and other small RNAs
The field of next-generation sequencing (NGS) has observed change at a rapid pace, with exponential gains in both data generation and scientific discoveries. The power of NGS lends itself to the examination of a variety of informationally dense biological materials, including DNA, RNA, miRNA, and other small RNAs. Sequencing of miRNA and other small RNAs, approximately 20-30 nucleotides in length, has provided key insights into understanding their biological functions, namely regulating gene expression and RNA silencing (see review, Gebert and MacRae, 2018). Additionally, studies have also identified and highlighted the importance of miRNAs as key biomarkers, novel therapeutic targets and how misregulation of miRNAs can result in the onset of many diseases and cancer (Witwer and Halushka, 2016).
SomaGenics RealSeq®-AC NGS RNA library preparation
Traditional NGS methodologies for investigating miRNA library preparations typically incorporate several key manipulations that are executed in sequential order. A simplified schematic of a NGS workflow for miRNAs may include but are not limited to the following steps: miRNA/adapter ligation, reverse transcription of the miRNA to cDNA coupled with PCR amplification (RT-PCR), library generation and analysis (Figure 1). Interestingly, there is growing evidence that suggests use of two sequencing adapters, flanking the 5’ and 3’ ends of the miRNA during preparation of the sequencing library, may result in deviations in the true abundance of each miRNA sequence (Fuchs et al., 2015). These deviations prove to be problematic as they result in significantly underrepresenting the quantity of many miRNAs or excluding them altogether. Compatible with Illumina/Solexa sequencing platform, RealSeq®-AC uses a single adapter that is ligated onto each miRNA sequence and provides increased efficiency at this step compared to alternate two-adapter ligation methods.
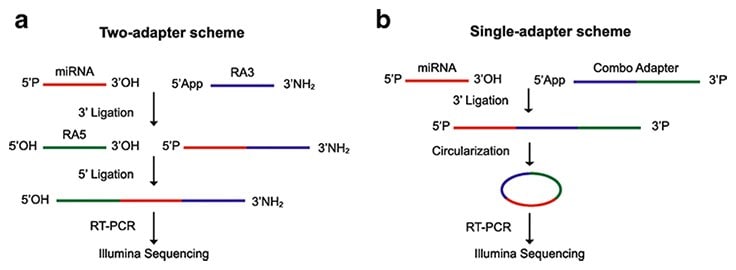
Figure 1.Comparison of two-adapter or single-adapter schemes
Source: Genome Biology Biomed Central
Introduction of RNA sequencing bias
Providing several advantages over traditional microarray techniques, including a better signal to noise ratio, NGS has proven to be a powerful method for sequencing RNA. RNA library preparation, reverse transcription, and PCR amplification are all critical stages within the NGS workflow; however, sequencing bias is often introduced to varying degrees during these stages. During RNA library preparation, small adapter ligands are incorporated onto the flanking regions of each RNA sequence. Several studies have reviewed the varying methods and technologies that are currently used during the formation of the miRNA-adapter ligand product and suggest that this step may introduce significant sequence bias (Dard-Dascot et al., 2018)
Reducing incorporation bias with RealSeq®-AC
As miRNA and small RNA NGS technology matures, researchers have identified unique challenges to accurately identify and quantify each type of RNA. Various strategies have been developed to remove bias that may arise from the sequence itself and potential biochemical modifications that may obscure the true abundance of various RNAs from a biological sample. For example, the 2’-O-methyl (2’-OMe) modification, present on the 3’ terminal nucleotide of specific populations of RNAs, varies in abundance depending on the sample source and reduces adapter ligation efficiency. The RealSeq®-AC gel-free protocol mitigates these bias-inducing factors with the use of a single standard sequence RealSeq® adapter that is ligated onto the 3’ end of each RNA sequence. Blocking and removal of unligated adapters, followed by intramolecular circularization, results in increased adapter ligation efficiencies compared to two-adapter methods (Figure 2). Studies have shown that not only is sequencing bias reduced, but also an increased number of small RNAs are identified compared to competitor RNA preparation kits (Barberán-Soler et al., 2018). Providing assurance of accurate reads for your precious samples, RealSeq®-AC libraries can be efficiently generated for RNA input ranging from 1-1000 ng of total RNA from cells or tissues.
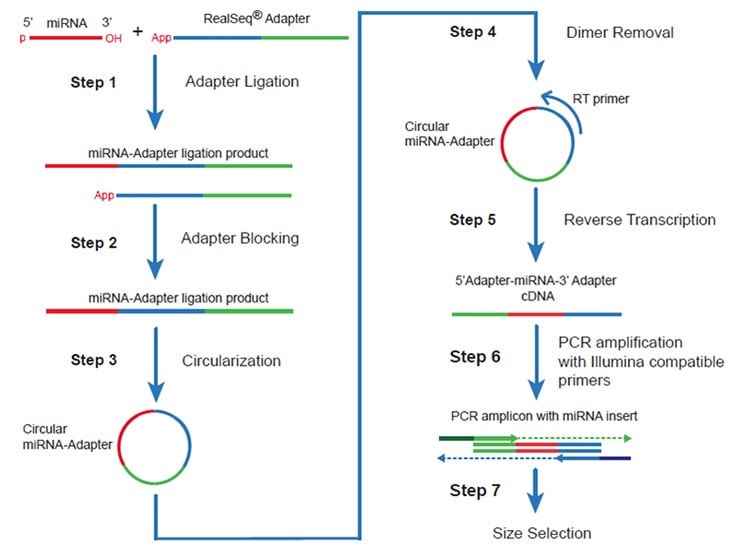
Figure 2.RealSeq®-AC RNA NGS library preparation and workflow
RNA library preparation support
Assisting you throughout your RNA preparation workflow, we offer both the technical and product support to drive your next discovery.
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?