ChIC and CUT&RUN Data Analysis Tool Overview
ChIP-seq is a powerful tool to investigate DNA-protein interactions. It can be used to map chromosomal locations of transcription factors (TFs), histones modifications, chromosomal DNA-binding enzymes, chaperones, or nucleosomes. Over the years, the fundamentals of Chromatin immunoprecipitation (ChIP)have remained unchanged. Issues remain with high background that limits sensitivity, requirements for large numbers of cells, and artifacts resulting from cross-linking and solubilization. Chromatin immunocleavage (ChIC) is an alternative important technique that overcomes the challenges and inefficiencies mentioned above. Based on the ChIC technique, researchers have made further advancements that make the ChIC technique applicable to genome-wide profiling with simplified handling requirements and is refered to as Cleavage Under Targets and Release Using Nuclease (CUT&RUN) 2. We developed three ChIC/CUT&RUN ChIC kits based on ChIC and CUT&RUN techniques, with an addition of pG-MN or pA-MN/pG-MN protein cocktail to provide more options for antibody selection (Cat. #: CHR100-102).
Even though the ChIC/CUT&RUN method has made significant improvements on the experimental procedure, the resulting sequencing data process and analysis remain the same as ChIP-seq. Therefore, we developed a data processing and analysis tool that is suitable for the three techniques mentioned above, namely ChIP-seq, ChIC and CUT&RUN. Currently, this tool is only available to our customers who purchase a ChIC/CUT&RUN kit. This data analysis tool provides a comprehensive analysis on ChIP-seq experimental results. The tool is organized into three modules including file management, data analysis and the results report (Figure 1).
Within the file management module, various functions allow users to load their NGS files, barcoded or un-barcoded that were previously generated on the Illumina platform. Associated functions include read file quality control (QC), visualization, a default function for unzip, and read pre-process to meet QC requirements. Each account is given 50GB storage disk space, while max file size is allowed for uploading 6G.
The data analysis modules provide three main functions, including peak calling, TF binding profile, and differentially binding analysis. Factors under consideration for data analysis include: peak type (narrow peak and broad peak), barcoded or un-barcoded samples, replicate number, and presence or absence of a control. Further annotation for each analysis function was provided under different categories ranging from binding site distribution, pathway/GO enrichment and Venn diagram. In our current version we mainly consider human (hg38, hg19), mouse (mm10, mm9), and Rat (rn6, rn4).
In the results module, the data for peak calling, motif discovery, differentially binding, and their annotation are all displayed by clicking their respective result panel. Peak calling results are provided for each individual sample and the consensus results for samples with >1 replicate. The results can be downloaded and visualized by IGV genome browser. With the TF binding profile function, the user can identify DNA motifs with selected top binding sites and the identified top DNA binding motifs are further compared with the Jaspar database. Comparative binding analysis identifies differential binding between different samples/treatments, where number of replicates is associated with the method that was used. Importantly, the annotation results for each type of analysis can be downloaded and visualized as a table and plot. For more information about the tool, please view the FAQs.
Your order ID can be found on the packing slip, the order confirmation or retrieved by calling your local Customer Service.
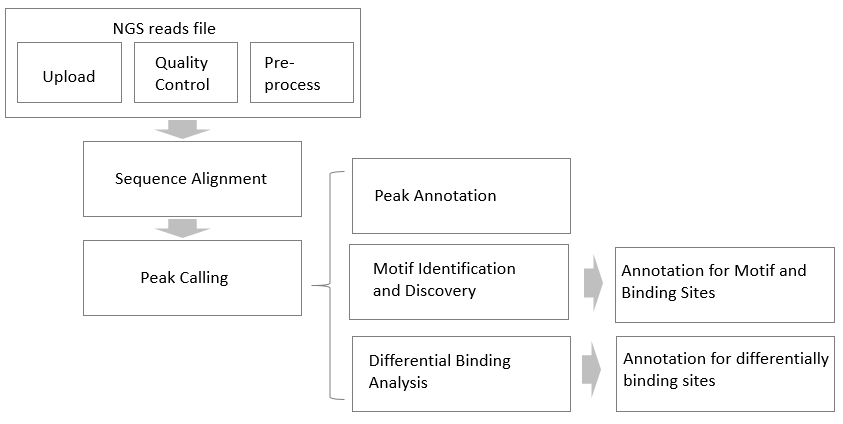
Figure 1.Data analysis workflow
References
To continue reading please sign in or create an account.
Don't Have An Account?