Digoxigenin (DIG) Labeling Methods
Section Overview
- Digoxigenin (DIG) Labeling and Anti-DIG Antibody
- DIG DNA Labeling by PCR
- DIG Random Primed DNA Labeling
- Nick Translation Labeling of dsDNA for In Situ Probes
- Transcriptional Labeling of RNA Probes
- DIG Oligonucleotide 5’ End-Labeling, 3’ End-Labeling, and 3’ Tail Labeling
- Estimation of Probe Yield by the Direct Detection Procedure
- DIG-Related Downloads and Resources
Digoxigenin (DIG) Labeling and Anti-DIG Antibody
The DIG System is the nonradioactive technology of choice to label and detect nucleic acids for multiple applications. The system is based on a steroid isolated from digitalis plants (Digitalis purpurea and Digitalis lanata). These plants are the only natural source of digoxigenin, so the anti-DIG antibody does not bind to other biological material, ensuring specific labeling. Due to this high specificity, less material is needed compared to radioactive labeling making the DIG system ideal for nucleic hybridization analysis. Immobilized nucleic acids are hybridized with a DIG-labeled probe and subsequent detection is performed using high affinity Anti-Digoxigenin antibodies, coupled either to alkaline phosphatase (AP), horseradish peroxidase (HRP), fluorescein or rhodamine for colorimetric, and chemiluminescent or fluorescent detection.
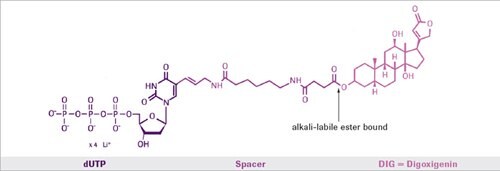
Figure 1.Example detecting DIG-labeled nucleic acids using chemiluminescence substrates.

DIG DNA Labeling by PCR
PCR labeling is the preferred method for preparing DIG-labeled probes when the template is available in only limited amounts, is partially purified, or is very short. It requires less optimization than other methods and produces a high yield of labeled probe. In PCR labeling, a thermostable polymerase incorporates DIG-dUTP as it amplifies a specific region of the template DNA. The result is a highly labeled, specific, and extremely sensitive hybridization probe.
Reaction Principle
During a standard PCR reaction, Digoxigenin-11-dUTP is incorporated into newly synthesized DNA. The only prerequisite is that some sequence information of the target sequence is needed in order to synthesize the appropriate primers. The nonradioactive DIG system uses digoxigenin, a steroid hapten, to label DNA, RNA, or oligonucleotides for hybridization, and subsequent color or luminescence detection. The digoxigenin is coupled to dUTP via an alkali-labile ester bond. The labeled dUTP can be easily incorporated by enzymatic nucleic acid synthesis using DNA polymerases. The combination of nonradioactive labeling with PCR is a powerful tool for the analysis of PCR products, and for the preparation of labeled probes from small amounts of a respective target sequence.
DIG DNA Labeling by PCR Features and Benefits
PCR Conditions
- Optimize PCR amplification parameters (cycling conditions, template concentration, primer sequence, and primer concentration) for each template and primer set in the absence of DIGdUTP before attempting incorporation of DIG.
Template
- For best results, use cloned inserts as template. Genomic DNA can be more difficult to use.
- Template concentration is crucial to successful production of specific probes.
Labeling
The PCR DIG Probe Synthesis Kit requires less optimization than most labeling methods, because it contains the Expand High Fidelity Enzyme Blend. This enzyme blend benefits include:
- Can efficiently use GC-rich regions as template
- For most templates, requires no optimization of MgCl2 concentration and labeling reactions will work with the standard concentrations of 1.5 mM MgCl2
DIG Random Primed DNA Labeling
The method of "random primed" DNA labeling is based on the hybridization of a mixture of all possible hexanucleotides to the DNA template. All sequence combinations are represented in the hexanucleotide primer mixture, which leads to binding of primer to the template DNA in a statistic manner. Thus, an equal degree of labeling along the entire length of the template DNA is guaranteed. The complementary strand is synthesized from the 3´ OH termini of the random hexanucleotide primer using Klenow enzyme, labeling grade. Modified deoxyribonucleoside triphosphates ([32P]-, [35S]-, [3H]-, [125I]-, digoxigenin or biotinlabeled) present in the reaction are incorporated into the newly synthesized complementary DNA strand.
These labeled probes are especially suitable for single copy gene detection on genomic Southern blots, screening recombinant libraries, dot/slot blots, and northern blots. Since each primer has a different six-base sequence, the labeled probe product will be a collection of fragments of variable length. Thus, the labeled probe will appear as a smear, rather than a unique band on a gel. The size distribution of the labeled probe depends on the length of the original template.
Reaction Principle
In random primed labeling, Klenow enzyme copies DNA template in the presence of hexamer primers and alkalilabile DIG-11-dUTP. On average, the enzyme inserts one DIG moiety in every stretch of 20-25 nucleotides. The resulting labeled product is a homogeneously labeled, sensitive hybridization probe capable of detecting as little as 0.10 – 0.03 pg target DNA.
Nick Translation Labeling of dsDNA for in situ Probes
The nick translation method is based on the ability of DNase I to introduce randomly distributed nicks into DNA at low enzyme concentrations in the presence of Mg2+. E. coli DNA polymerase I synthesizes DNA complementary to the intact strand in a 5´ - 3´ direction using the 3´-OH termini of the nick as a primer. The 5´ - 3´ exonucleolytic activity of DNA Polymerase I simultaneously removes nucleotides in the direction of synthesis. The polymerase activity sequentially replaces the removed nucleotides with isotope-labeled or hapten-labeled deoxyribonucleoside triphosphates. At low temperature (+15 °C), the unlabeled DNA in the reaction is thus replaced by newly synthesized labeled DNA. For in situ hybridization procedures, the length of the labeled fragments obtained from this procedure should be about 200-500 bases.
Transcriptional Labeling of RNA Probes
For some applications, DIG-labeled RNA is a more effective hybridization probe than DIG-labeled DNA. For example, DIG-labeled RNA probes can detect rare mRNAs in nanogram amounts of total RNA. These labeled RNA probes are generated by in vitro transcription from a DNA template. In the RNA transcription method, DNA is cloned into the multiple cloning site of a transcription vector between promoters for different RNA polymerases (such as T7, SP6, or T3 RNA polymerase). The template is then linearized by cleavage of the vector at a unique site (near the insert). An RNA polymerase transcribes the insert DNA into an antisense RNA copy in the presence of a mixture of ribonucleotides (including DIG-UTP). During the reaction, the DNA can be transcribed many times (up to a hundredfold) to generate a large amount of full-length DIG-labeled RNA copies (10-20 μg RNA from 1 μg DNA in a standard reaction). DIG is incorporated into the RNA at approximately every 25-30 nucleotides.
Reaction Principle
The DNA template to be transcribed is cloned into the polylinker site of an appropriate transcription vector, which contains promoters for SP6 and or T3 and T7 RNA polymerases. After linearization at a suitable site, RNA is transcribed in the presence of DIG-11-UTP. Under standard conditions, approximately 10 μg of full-length DIG-labeled RNA are transcribed from 1 μg of template.
The following tips are critical for successful RNA probe labeling:
RNases
RNases are ubiquitous and do not require any cofactors for activity. If you want to be successful, take all possible precautions to prevent RNase contamination. For instance:
- It is recommended to use disposable plasticware, oven-baked glassware, or plasticware that has been decontaminated with RNase ZAP or similar reagents.
- Prepare all solutions with water that has been treated with diethyl-pyrocarbonate (DEPC) or dimethyldicarbonate (DMDC) and autoclave the solutions.
- Wear gloves throughout the procedure.
- Labeling efficiency depends greatly on the purity of the DNA template. Template should be highly purified.
- The final template must be linearized, phenol/chloroform extracted, and ethanol precipitated.
Template Sequence
- Some primer and/or polylinker regions in DNA templates are homologous to portions of the ribosomal 28s and 18s RNA sequences. Therefore, labeled probes may generate specific, but unwanted signals in samples that contain these prominent RNAs. To minimize this effect, remove as much of the polylinker sequences from your template as possible.
- If you use PCR to make the DNA template, the product of the Expand High Fidelity reaction contains some fragments with a single 3´ A overhang. This overhang may produce wraparound products in the transcriptional labeling reaction.
Template Length
- Optimal template length is approximately 1 kb
- Minimum length should be at least 200 bp
Storage of Probe
- For long term stability, RNA probes should be aliquoted and stored at -20 °C or -70 °C
- DIG-labeled RNA probes are stable for at least 1 year at -20 °C or -70 °C in ethanol
Probe Sensitivity
- To quickly determine the sensitivity of a DIG-labeled antisense RNA probe, prepare the corresponding sense RNA (unlabeled) by in vitro transcription. Then use the purified sense transcript at varying concentrations as target on a northern blot. From the result of the blot you can easily determine the lowest amount of target (sense transcript) that can be detected by labeled probe (antisense transcript).
DIG Oligonucleotide 5’ End-Labeling, 3’ End-Labeling, and 3’ Tail Labeling
For some applications, such as in situ hybridization, a DIG-labeled synthetic oligonucleotide is the best hybridization probe. In addition to in situ hybridizations, DIG-labeled oligonucleotides may be used as hybridization probes in numerous applications, including dot/slot blots, library screening, detection of repeated gene sequences on Southern blots, and detection of abundant mRNAs on northern blots.
Several methods are available for DIG-labeling of oligonucleotides and are summarized below.
Oligonucleotide 5´ End-labeling with DIG-NHS-Ester
Oligonucleotide 3´ End-labeling with DIG-ddUTP
Adding a 3´ Tail of DIG-dUTP and dATP (approximately 40 - 50 residues)
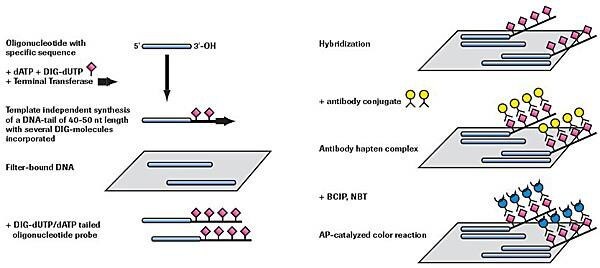
Figure 2.Nonradioactive oligonucleotide tailing and detection.
Estimation of Probe Yield by the Direct Detection Procedure
To add the correct amount of probe to a hybridization, you must first determine the amount of DIG-labeled probe produced in the labeling reaction. The direct detection procedure given here compares the amount of DIG label in a series of dilutions prepared from the labeled probe with a known concentration of a DIG-labeled control nucleic acid.
Note: If you label a DNA probe by PCR, you do not need to perform a direct detection to evaluate the yield.
For PCR-labeled probes, use the gel electrophoresis evaluation method.
The direct detection involves the following steps:
- Preparing serial dilutions of labeled probe and spotting them on a nylon membrane (time required: 15 minutes)
- Detecting DIG in spots with chemiluminescence; time required (2-2.5 hours)
DIG-Related Downloads and Resources
Need help? Check out our DIG product selection guide.
For more information, view the following brochures:
Detailed technical information can be found in the DIG Manuals:
- DIG Application Manual for Filter Hybridization
- DIG Application Manual for Nonradioactive In Situ Hybridization, 4th edition
Learn how to handle the DIG System in our Technical Tips:
For life science research only. Not for use in diagnostic procedures.
Related Products
To continue reading please sign in or create an account.
Don't Have An Account?