Universal ProbeLibrary System Performance Data
Comparison to Other Assay Formats
Comparison to HybProbe Assays on the LightCycler® 2.0 Instrument
Comparison to SYBR Green I Assays
Comparison to Other Commercially Available Predesigned Custom Assays
Comparison to a Commercially Available Hydrolysis Probe Assay
Application for Studying Gene Knockdown Effects By RT-PCR
Overview
Design Intron-Spanning Assays that Work the First Time
When you need primers and probes for your real-time qPCR assay, don't waste time using a cumbersome trial-and-error design process. Simply go to the Assay Design Center and use the highly sophisticated, extensively tested features of the ProbeFinder software. This software can suggest several assays for each target, ranked according to their probability of success. If results obtained with the highest ranked assay are not satisfactory, you can simply try the second assay suggested by the software; you don't need to waste time optimizing the first assay.
When the software was used to design assays for 175 of the most frequently cited human RefSeq transcripts, 84% of the highest-ranked assays suggested by the software detected the transcript. Using the second suggested assays, another 12% of the transcripts could be detected successfully.
Coverage Rate
Universal ProbeLibrary enables the fast and easy design of intron-spanning assays for a large number of organisms. The ProbeFinder software will design assays with high coverage rates for the listed genomes:
Universal ProbeLibrary Assays and the Appropriate Master Mix on Your Real-Time PCR Instrument
The Universal ProbeLibrary assays are compatible with all real-time PCR instruments capable of detecting fluorescein, FITC, FAM and/or SYBR Green I. The Universal ProbeLibrary assays have been used successfully on the LightCycler® Carousel-Based Instruments, the LightCycler® 480 System, and other real-time PCR instruments from several suppliers (please also see the Performance and Applications section).
Using the appropriate real-time PCR master mix for your real-time PCR instrument can make a difference:
Universal ProbeLibrary Assays Perform Comparable to Other Assay Formats
Comparable results in terms of linearity are obtained with Universal ProbeLibrary probes, HybProbe probes or SYBR Green I.
The three different types of assays were used to detect three human mRNA targets (IL-8, IFN-γ and IL1β). Each assay was performed on four different concentrations (undiluted, 1:100 dilution, 1:1,000 dilution and 1:10,000 dilution) of the same human template cDNA pool. Real-time PCR assays were performed in duplicate on a LightCycler® 1.2 Instrument.

Figure 1.Linear regression analysis (i.e., the logarithm of template concentration versus crossing point cycle values (CP)) for SYBR Green I (green), Universal ProbeLibrary (blue), and HybProbe assays (magenta). All assays were quite linear over four orders of magnitude.
Data kindly provided by Dr. Thomas Giese, Institute of Immunology, University of Heidelberg, Germany
Universal ProbeLibrary Assays Perform Comparable to HybProbe Assays on the LightCycler® 2.0 Instrument
A real-time PCR assay for β2-Microglobulin (β2M) was performed on serial dilutions of the same in vitro transcribed human RNA (106, 105, 104, 103, 102, 101 copies, no template control (NTC)). Each assay was performed in triplicate on a LightCycler® 2.0 Instrument. The LightCycler® FastStart DNA MasterPLUS HybProbe was used for the HybProbe assay. The LightCycler®TaqMan® Master was used for the Universal ProbeLibrary assay.

Figure 2.Amplification of β2M with Universal ProbeLibrary probes (left) and with HybProbe probes (right).
Universal ProbeLibrary Assay Performance Compares to SYBR Green I Assays, but no Primer-Dimers are Detected
Universal ProbeLibrary offers the flexibility of SYBR Green I, by being independent from designing and ordering of fluorescence labeled probes. In addition, Universal ProbeLibrary assays provide a specificity which is comparable to commonly used probe formats, such as hydrolysis probes or HybProbe probes.
To demonstrate this, SYBR Green I assays were directly compared to Universal ProbeLibrary assays in terms of specificity, determined by use of a no-template-control (NTC). In the figure below it is clearly shown that amplification of the NTC is not detectable in an Universal ProbeLibrary assay, whereas a strong amplification signal is always detected in SYBR Green I

Figure 3.Real-time RT-PCR assays with a dilution series for GAPDH cDNA on a LightCycler® 2.0 Instrument. A: Amplification curve of SYBR Green I assay; B: Melting curve analysis of the SYBR Green I assay; C: Amplification curve of Universal ProbeLibrary assay; NTC = No template control.
Data is kindly provided by Amy Jassen, Ph.D., New England Primate Research Center, Harvard Medical School
Performance of Universal ProbeLibrary Assays is Directly Comparable to Other Commercially Available Predesigned Custom Assays, run on a Competitor Block-Based Real-Time PCR Instrument
RNAs were isolated from cells expressing the respective target genes and transcribed with the Transcriptor First Strand cDNA Synthesis Kit. The assays were run on a competitor block-based real-time PCR instrument. Universal ProbeLibrary assays were performed with the FastStart TaqMan® Probe Master, competitor assays with the recommended competitor PCR Master.

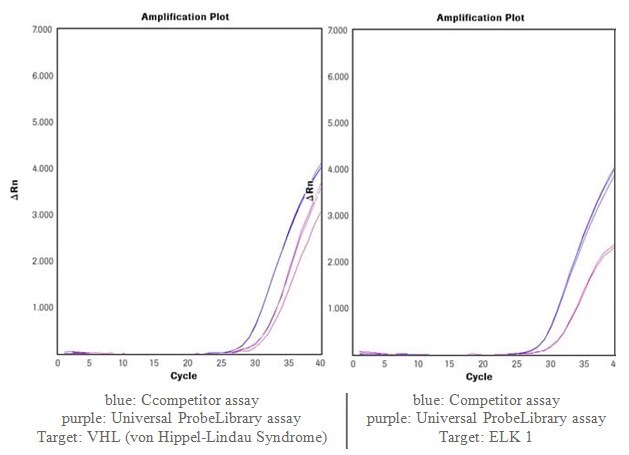
Figure 4.Comparison of Universal ProbeLibrary assays to other commercially available predesigned custom assays. (Data is kindly provided by PD Dr. S. Wiemann, German Cancer Research Center - DKFZ, Germany)
Comparison of a Commercially Available Hydrolysis Probe Assay to a Universal ProbeLibrary Assay with two Different Primer and Probe Concentrations Run on the LightCycler® 480 Instrument
GAPDH was amplified from dilutions of a qPCR human reference cDNA (BD Biosciences) performing a Universal ProbeLibrary assay with probe # 60 and appropriate primers and a commercially available competitor hydrolysis probe assay for GAPDH.
The Universal ProbeLibrary assay was performed with the recommended standard concentrations of primers and probe (200 nM primers and 100 nM probe) and with the same (elevated) concentrations of primers and probe (900 nM primers and 250 nM probe) as used in the competitor assay. Both type of assays were run on a LightCycler® 480 Instrument. A standard run protocol was performed and for both assays the LightCycler® 480 Probes Master was used.
All three assays perform with optimal PCR efficiency of approx. E = 2.
When performing the Universal ProbeLibrary assay with the recommended standard concentration of probe (100 nM) a lower fluorescent signal is generated. With elevated probe concentration (250 nM) the signal height is the same as that of the competitor assay.
The Universal ProbeLibrary assay for GAPDH creates slightly earlier crossing points than the competitor assay.
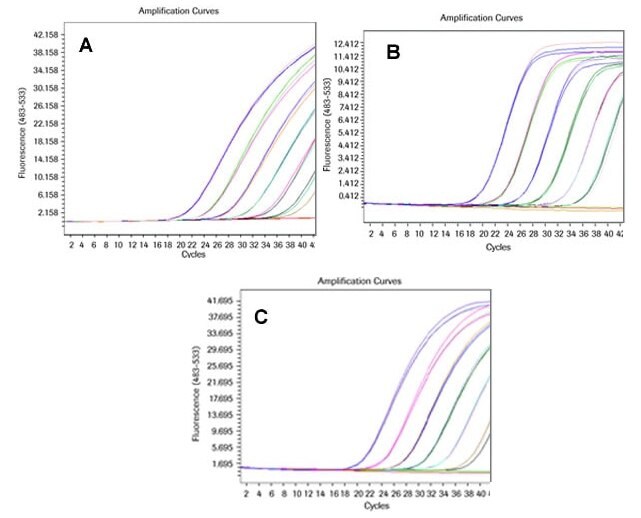
Figure 5.Real-time RT-PCR assays with a dilution series of commercially available human reference cDNA for GAPDH on a LightCycler® 480 Instrument.
A: Amplification curve of a commercially available pre-validated hydrolysis probe assay.
B: Amplification curve of a Universal ProbeLibrary assay with standard primer (200 nM) and probe (100 nM) concentrations.
C: Amplification curve of a Universal ProbeLibrary assay with elevated primer (900 nM) and probe (250 nM) concentrations.
Application of Universal ProbeLibrary for Studying Gene Knockdown Effects by RT-PCR
RNAi has become a widely used approach to studying the phenotypic effects of knocked-down components of many pathways implicated in physiology and disease. Stringent quality-control procedures to monitor RNAi experiments are essential. Requirements for such a methodology are (a) high flexibility in assay design if many different genes need to be examined, (b) precision in assessing gene expression levels, and (c) ability to measure expression levels of multiple genes in parallel.
A versatile design of RT-PCR assays and the necessary experimental optimization remain a significant technical challenge. To perform large-scale RT-PCR experiments, it would be desirable to combine the flexibility of an "out-of-the-box" SYBR Green I assay with the specificity of hybridization probes. We used signal transduction systems in Drosophila as a model and developed assays that measure the mRNA concentration of target genes using the Drosophila Universal ProbeLibrary. These assays allowed us to predict models of pathway topologies.
We first designed RT-PCR assays for a series of target genes. Assays designed included Mtk, AttA, CecA2, and Puc that respond to both IMD branches. The ProbeFinder software was used to identify primer pairs and corresponding probes (Table 1). As a first step, we tested the performance of the assays by determining primer efficiencies using a serial dilution of target gene cDNA. As shown in Figure 6a, RP49 primers performed in a range between 0.052 pg and 52 pg of cDNA, with a primer efficiency of 2.0. Similar results were obtained for all other designed RT-PCR assays which had an expected efficiency of 1.9 -2.0 (data not shown).

To continue reading please sign in or create an account.
Don't Have An Account?