Microfluidics for Nanoencapsulation of Nucleic Acids
Introduction
In contrast to lipid nanoparticles (LNPs), which are commonly actively loaded with nucleic acids, polycations such as polyamines self-assemble with polyanionic nucleic acids into so-called polyelectrolyte complexes (short polyplexes) due to electrostatic attraction. Their disadvantage compared to LNPs, is their less defined morphology and composition, which has been reported to lead to polydisperse particle size distribution and poorly reproducible formulations.1 By microfluidic assembly of nucleic acid/carrier complexes, both disadvantages can be addressed efficiently for formulating defined and reproducible RNA nanocarriers.2 For more information on microfluidics for the preparation of LNPs, we recommend Reference 3. Amongst polyamines used for complexation, polyethylenimine (PEI) and poly-L-lysine (PLL), as well as their derivatives, are most widely used for preclinical applications. Additionally, polyamidoamine (PAMAM) dendrimers, poly(2-(dimethylamino)ethyl methacrylate (PDMAEMA), or polyamines of natural origin, such as chitosan, cationic gelatin or pullulan, and arginine-rich peptides have been described for polyplex formation.4 Polyamines, which have been chosen in various studies for microfluidic assembly of nucleic acid polyplexes, are depicted in Figure 1.
Section Overview

Figure 1.Cationic polymeric carriers used for microfluidic mixing with nucleic acids. For poly(amidoamine) (PAMAM) dendrimers, PAMAM generation 2 is shown. For more information on PAMAM dendrimers, we refer to Reference 5.
Microfluidic Mixing Using Flow Focusing
The first microfluidic systems with cationic carriers for encapsulating nucleic acids were used in 2009 to encapsulate plasmid DNA (pDNA) in 25 kg/mol hyperbranched PEI. Microfluidic mixing was performed to prevent large particle size distributions of DNA/PEI complexes caused by conventional bulk mixing and to improve general particle characteristics, cell viabilities, and transgene expression. A simple poly(methyl methacrylate) microfluidic hydrodynamic focusing device (MF) was used at flow rates of 10 and 50 µL/min for the middle (DNA solution) or side channels (PEI solution), respectively. The DNA stream was hydrodynamically focused into a narrowly focused stream by the two PEI side streams (Figure 2).6
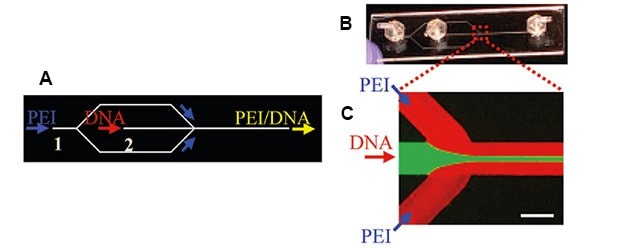
Figure 2.A and B) Microfluidic hydrodynamic focusing device with two inlets and one outlet for the preparation of pDNA/PEI complexes. C) Fluorescence micrograph of the flow pattern using fluorescein dye and Rhodamine-labeled PEI. Reproduced with permission from reference 6, copyright 2009, American Chemical Society.
A similar simple microfluidic device was used to study the DNA polyplex self-assembly under laminar flow in real time using quantum-dot mediated fluorescence resonance energy transfer (QD-FRET) studies. pDNA was labeled with quantum dots as the FRET-donor, and Cy5-dye-conjugated chitosan was used as the biodegradable, non-toxic, cationic carrier and FRET acceptor. Due to the preservation of a laminar flow after combining both solutions in a T-junction, mixing solely took place at the interface of both streams enabling precise self-assembly kinetics monitored by FRET. Detection of Cy5 emission confirmed successful DNA/chitosan polyplex formation. Emission was detected right after combining both flows in the T-junction, showing immediate DNA/carrier interactions with increasing polyplex concentrations over time.7
Three-dimensional hydrodynamic flow focusing was chosen by Quinones-Hinojosa et al. to produce poly(β-amino ester)/DNA polyplexes. The continuous flow assembly of these polyplexes was performed at a rate of 100 mg/h using a high flow rate of 45 ml/h. To overcome problems of long-term storage of polyplexes or polyesters in aqueous solutions, nanocomplexes were lyophilized. After three months of storage at -20 °C, these particles showed similar transfection efficiencies in glioblastoma cells. Still, in melanoma and breast cancer cells, transgene expression was only comparable to freshly prepared polyplexes when the particles were prepared using microfluidics. By nanoparticle tracking analysis of polyplexes loaded with Cy3-labeled plasmids, the authors attempted to determine nanoparticle concentration and the number of plasmids per particle.8
Microfluidic Mixing Chips with Loops
To increase interactions between polymers and nucleic acid solutions in the microfluidic channels, active mixers are equipped with baffles that induce controlled turbulent flow rather than laminar flow. In one example, Kissel et al. investigated the formation of PEI/pDNA polyplexes using a microfluidic device and polyplexes with modified PEI (hyperbranched, 25 kg/mol and 5 kg/mol) and siRNA or mRNA as nucleic acids. Polyplexes were prepared in a microfluidic lab-on-a-chip assembly using different flow rates. PEI and nucleic acid solutions were pumped separately into a mixing chip composed of different geometries with varying mixing lengths and designs. Mixing behaviors in the different chips were evaluated with trypan blue and 5(6)-carboxyfluorescein solutions under a microscope. As some designs showed a laminar flow of both solutions even after traversing the Y-junction without any subsequent mixing, a loop was necessary following the Y-part to facilitate the mixing of PEI and DNA to form small and uniform polyplexes. The particle sizes of PEI/pDNA polyplexes mainly depended on the ratio between the cationic carrier and nucleic acid (N/P ratio) with uniform sizes at N/P = 5–20 and independent from concentrations or flow rates.9 Further on, the group used a lower-molecular weight fraction of 25 kg/mol PEI, 5kg/mol PEI and Transferrin-PEI (5 kg/mol) for specific targeting approaches in combination with pDNA, siRNA, and mRNA. For pDNA, polyplex sizes were similar regardless of the preparation method, but polydispersities were lower for microfluidic mixing compared to pipetting. The same microfluidic mixing method was transferable to other nucleic acids since similar sized polyplexes with these polymers and siRNA or mRNA were formed; generally, higher polydispersities were observed when using siRNA.9
The groups of Kissel and Merkel evaluated a more sophisticated PEI copolymer. An amphiphilic water-insoluble triblock copolymer consisting of a hydrophilic poly(ethylene glycol), hydrophobic poly(caprolactone), and 2.5 kg/mol linear PEI (PEG-PCL-PEI) was used as an siRNA carrier.10 Microfluidic mixing led to the smallest, most uniform, and most stable particles compared to all other tested techniques when using a solution of pre-formed nanocarrier prepared via solvent-displacement technique and a nucleic acid solution using the same microfluidic setup that was most efficient in their previous study (vide supra). In addition, slightly higher transfection efficiencies were observed in SKOV3 cells for these polyplexes, in which the knockdown was increased from 24.5% (polyplexes via pipetting) to 34.8%.10 Merkel et al. further developed these studies using a water-soluble PEG-PCL-PEI polymer of hyperbranched 25 kg/mol PEI. A Dolomite micromixer chip was used in which a polymer and an siRNA solution were individually pumped into the chip with a flow rate of 0.5 mL/min (Figure 3). At this lowest flow rate of the three tested ones, polyplexes were only half the size of those prepared by bulk mixing with a narrower size distribution. While the smaller size did not result in any benefits regarding in vitro transfection, it was clearly shown that these polyplexes were more efficiently taken up in the lung parenchyma after intratracheal in vivo administration, which was discussed as a result of less efficient clearance by macrophages in the lung.2 The same group showed that this setup efficiently sensitized lung cancer cells to cisplatin using siRNA targeted to ERCC1 and XPF. After downregulating two of the most efficient pathways that repair platinum-induced DNA damage by 80–90% in p53 wildtype lung cancer cells, their IC50 sensitivity toward cisplatin was decreased by factor 2.11

Figure 3.Schematic drawing of microfluidic mixing of PEI and siRNA using a Dolomite Micromixer Chip. Reproduced with permission from reference 2, copyright 2017, IOP Publishing.
A more complex system in which cationic 2.5 kg/mol PEI-β-cyclodextrin (PEI-CD), hyaluronic acid-adamantane (HA-Ad), and pDNA were mixed in a commercial Chemtrix microreactor was developed by Thompson et al. using a layer-by-layer approach (Figure 4). PEI-CD and pDNA were assembled through two inlets at N/P ratios of 10, 20, or 30 and pDNA flow rates of 5, 10, or 20 µL/min. After self-assembly, HA-Ad was added in a separate inlet at the end of the mixing chip. HA-Ad was used due to its water solubility, CD44 targeting capability, and biocompatibility to serve as a protective shell. Higher flow rates led to smaller particles with narrower size distributions and lower Zeta-potentials compared to a similar bulk mixing procedure. However, polyplex stability studies with PicoGreen indicated that polyplexes prepared by flow mixing were less stable in the presence of heparin as a competing polyanion and more susceptible to disassembly. The reduced stability in combination with reduced Zeta-potentials was hypothesized to cause decreased transfection efficacies for flow-mixed complexes on the one hand with decreased cytotoxicity on the other hand.12
In a similar two-step approach, Moffitt et al. studied the self-assembly of cationic poly(2-vinylpyridine)-block-poly(ε-caprolactone) (P2VP-PCL) with pDNA, followed by a second assembly with micellar poly(ethylene glycol)-block-poly(ε-caprolactone) (PEG-PCL) resulting in “polyplex-in-hydrophobic core” (PIHC) micelles. The microfluidic technique was solely used for the second assembly step to control the mixing of water and dioxane with a solution of the previously assembled P2VP-PCL/pDNA polyplex already combined with the PEG-PCL polymer.13
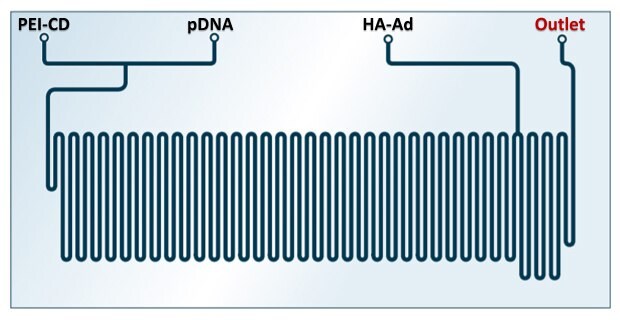
Figure 4.Chemtrix microreactor (Design 3023) with three inlets and one outlet for the formation of PEI-CD, pDNA polyplexes that are further assembled with HA-Ad. Reprinted with permission from reference 14, copyright 2022 Chemtrix BV.
Other Microfluidic Micromixer
Polyplexes with a 22 kg/mol linear PEI and pDNA were prepared with a micro-mixer at different speeds to investigate the impact of mixing speed on the polyplex characteristics. The up-scaled micromixer was composed of two syringe pumps pumping the PEI- and DNA-solution into a conventional T-shape connection, commonly used as a connector in HPLC analysis. Different speeds and plasmid concentrations were set to investigate the influence on polyplex formation. An increasing rate of mixing and a decrease in DNA concentration led to decreased and reproducible particle sizes (down to 50 nm) and exceptionally low polydispersities of 0.05. However, metabolic and transfection activities in vitro showed no differences between polyplexes prepared via up-scaled micro-mixing or more heterogeneous particles prepared by pipetting at a large volume.15
A more complex setup was investigated by Foged et al. in which three inlets were connected to a micromixer. One syringe with siRNA solution in the center inlet and two syringes with generation 4 PAMAM dendrimer in the outer inlets were pumped into a 3D printed micromixer to form polyplexes. The tailor-made micromixer was designed according to the desired flow pattern after performing simulation studies. Testing different flow rates, increased flow rates led to smaller particles. However, these sizes and polydispersities were not improved compared to bulk mixing at an N/P ratio of 20. Afterward, these polyplexes were transferred into microparticle-based powders via spray drying for inhalation therapy. Different saccharides were used during the spray drying process to obtain nanoembedded-microparticles (NEM). Several nanocomplex-to-matrix (N/M) ratios were tested with all excipients. Spheric particles with aerodynamic particle diameters of 5 µm with residual moisture contents between 0.2 (for mannitol) and 6 wt.% (for trehalose) were obtained. The viability of RAW264.7 cells was only slightly affected by the nanocomplexes at siRNA concentrations up to 400 nM. Cell uptake of NEM in the same cell line was verified via confocal microscopy using fluorescent siRNA and flow cytometry showing the highest cellular uptake for NEMs with a trehalose/inulin mixture of 50/50. Furthermore, this NEM induced an 87% inhibition of TNF-α expression in gene silencing experiments which was in the same range as freshly prepared nanocomplexes without excipient. The specific impact of microfluidic mixing alone on transfection efficacy was not investigated in this study.16
Droplet-based Approaches
By using the commercially available transfection agents jetPEI®, which is a 20 kg/mol linear polyethylenimine derivative, or Turbofect (poly(2-hydroxypropylenimine)) and pDNA in microfluidics-assisted confinement (MAC), Leong et al. were able to produce picolitre droplets consisting of well-defined nanocomplexes. The droplets were produced in a crossflow microfluidic droplet generator via competing flows of the continuous oil phase and disperse aqueous phase. Plasmid DNA, PEI, and buffer solutions were injected into an individual channel each via syringe pumps. DNA and carrier solutions were confined into individual droplets and were mixed in a serpentine channel to form nanocomplexes via electrostatic interactions (Figure 5). Buffer solution was used to avoid aggregation. Polyplexes formed via this method showed more monodisperse diameters with slightly more neutral Zeta-potentials and an increased nanocomplex stability compared to bulk mixing indicating optimal encapsulation of pDNA and resistance to aggregation. After incubation with HEK293 cells, polyplexes formed via microfluidics showed increased cell viability for both polymers and slightly better transfection when using Turbofect. It was shown that larger polyplexes resulting from bulk mixing partially showed higher uptake efficacies at early time points but leveled off. In contrast, smaller and more stable MAC-produced particles had a more consistent uptake.17 This observation is perfectly in line with the hypothesis that larger bulk particles sediment faster and may therefore result in efficient uptake,2 particularly at early time points,17 masking potential benefits of microfluidic mixing.15
The same group investigated the self-assembly of bioreducible poly(amido amines) with pDNA or mRNA in the same microfluidic device. Improved nanoparticle characteristics (smaller, more uniform, less aggregation) were observed. QD-FRET and flow cytometry were used to quantify the intracellular unpacking of polyplexes and DNA release. DNA-polyplexes produced by MAC showed increased colloidal and binding stability, and transfection was improved in various cell lines (HEK293, primary mouse embryonic fibroblasts, human mesenchymal stem cells, and HepG2 human hepatocellular carcinoma cells). These higher efficiencies were caused by more gradual release of the payload as monitored by QD-FRET.18

Figure 5.Microfluidic assembly of pDNA polyplexes in picolitre droplets using a crossflow type droplet generator. Reproduced with permission from reference 17, copyright 2011, American Chemical Society.
Chen et al. designed a similar droplet-based microfluidic device coupled to a dielectrophoresis (DEP) setup to directly sort polymer/pDNA nanocomplexes from the oil phase in a non-uniform electric field. As a cationic carrier, low molecular weight fatty acid-cyclodextrin-PEI (600 g/mol) was used, which already showed high transfection efficiency and low cytotoxicity in previous studies. DEP further decreased polydispersity and sizes of polyplexes compared to batch mixing/pipetting and on-chip mixing without DEP. Transfection efficacy, claimed to be increased by DEP mixing, was only assessed by microscopic counting of fluorescent cells, but no quantitative results were presented.19
Conclusion
Microfluidic mixing allows for controlled assembly of nucleic acid polyplexes, which in several accounts was reported to have beneficial effects on polyplex size and polydispersity. Regarding efficacy, the picture is not clear, which could result from most studies investigating only in vitro transfection. As pointed out, uptake kinetics depend on particle size in typical two-dimensional cell culture setups. Larger particles that sediment faster could mask potential benefits that smaller particles offer in the long run. In an in vivo study, clearance of larger particles by phagocytosing cells plays a more significant role; therefore, differences can be shown more clearly. Overall, microfluidic mixing conditions must be optimized for each mixing system and may also be affected by the materials and solvents used. The most significant advantage, however, is the ability to reproduce and scale up the production process.
Acknowledgment
F.A. is grateful for funding from the Federal Ministry of Education and Research (BMBF) and the Baden-Württemberg Ministry of Science, Research and Art as part of the Excellence Strategy of the German Federal and State Governments. O.M.M. acknowledges support from the European Research Council (ERC-2014-StG – 637830), from the Bavarian Research Foundation (AZ-1449-20C) and from the Center for NanoScience at LMU Munich.
NanoFabTx™ Microfluidic Device Kits
Poly(ethyleneimine) (PEI) for Gene Delivery
Chitosan for Gene Delivery
Microfluidic Mixing Chips and Droplet Generators
References
To continue reading please sign in or create an account.
Don't Have An Account?