Chart 1. End groups of different PEG macromers.
Andrea M. Kasko
Department of Bioengineering University of California, Los Angeles 410 Westwood Plaza, 5121 Eng V Los Angeles, CA 90095 United States
Progress in biotechnology fields such as tissue engineering and drug delivery is accompanied by an increasing demand for diverse functional biomaterials. For decades, research in polymeric biomaterials has focused on testing the biocompatibility of polymers developed for other applications and/or their processing (e.g., electrospinning, solvent casting/porogen leaching, 3D printing). More recently, researchers have shifted towards synthesizing materials specifically for biomedical uses, including synthetic proteins, glycomimetics and polymers compatible with aqueous media, along with chemical modification of naturally occurring polymers (e.g., to allow gelation or increased in vivo stability). In the past decade, polymer chemists have created a niche for designed biomaterials to use as cell scaffolds and to deliver therapeutic agents.
One class of biomaterials that has been the subject of intense research interest is hydrogels.1 Hydrogels are extensively investigated as two- and three-dimensional scaffolds for cells because they closely mimic the natural environment of cells, both chemically and physically.2 Hydrogels can be formed from synthetic (e.g., poly(ethylene glycol), poly (hydroxyethyl methacrylate)) and naturally occurring polymers (e.g., collagen, hyaluronan, heparin),1b and are useful 3D models of tissue culture due to their high water content and ability to form in the presence of cells, proteins and DNA. Depending on the reactivity of the constituent materials, gelation can be induced using pH3, temperature4, coulombic interactions, covalent bonding, non-covalent interactions5, or polymerization.
Poly(ethylene glycol) is a hydrophilic polymer that, when cross-linked into networks, can have a high water content. PEG is a suitable material for biological applications because it does not generally elicit an immune response.6 Since the 1970s, PEG has been used to modify therapeutic proteins and peptides to increase their solubility, lower their toxicity and to prolong their circulation half-life.7 In the late 1970s, researchers began to experiment with PEG hydrogels for cell culture. PEG hydrogels are chemically well-defined, and multiple chemistries can be used both for their formation and chemical modification.
PEG is easily synthesized by the living anionic ring-opening polymerization of ethylene oxide; well-defined (low polydispersity) PEGs with a range of molecular weights and a variety of end groups (e.g., alcohol, methyl ether, amine, N-hydroxysuccinimidyl (NHS) ester) are widely available.
In order to form a hydrogel, PEG must be cross-linked. Initially, PEG was cross-linked non-specifically using ionizing radiation.8 PEG hydrogels are now typically synthesized via covalent cross-linking of PEG macromers with reactive chain ends.
PEG macromers with reactive chain ends such as acrylate, methacrylate, allyl ether, maleimide, vinyl sulfone, NHS ester and vinyl ether groups (Chart 1) are easily synthesized from readily available starting materials. The alcohol chain ends of PEG can be esterified using acid chlorides (e.g., acryloyl chloride, methacryloyl chloride) in the presence of base. PEG chain ends can be etherified under basic conditions by reaction with alkyl halides such as 2-chloroethyl vinyl ether or allyl bromide. PEG divinyl sulfone is prepared by coupling PEG to a large excess of divinyl sulfone or by a multistep process to prepare chloroethyl sulfone chain ends that undergo basic elimination to form vinyl sulfone groups9.
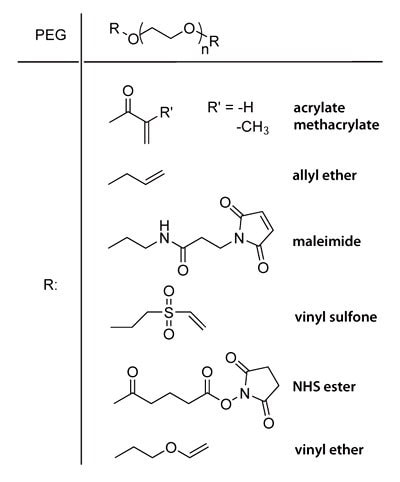
Chart 1. End groups of different PEG macromers.
Macromers can be homobifunctional or heterobifunctional. Homobifunctional macromers are typically used to form networks, while heterobifunctional macromers may be used to tether a therapeutic molecule into a hydrogel network.
The cross-linking mechanism to form hydrogels depends on the identity of the chain ends of PEG macromers. In most cases, cross-linking occurs when the reactive vinyl chain ends are polymerized, usually with a free radical initiator. For example, polymerization of macromers can be initiated using redox-generated radicals (e.g., ammonium persulfate and TEMED), or radicals generated with light (e.g., Irgacure® 651, λ=365 nM Scheme 1). Acrylate and methacrylate chain ends undergo chain polymerization. In step growth network formation, a multifunctional (f>2) cross-linker reacts with the PEG chain ends in a stoichiometric manner; alternatively, multifunctional PEGs (f>2) can be crosslinked with difunctional crosslinkers (Scheme 1). Acrylate, methacrylate, vinyl sulfone, maleimide, vinyl ether and allyl ether are all capable of step growth network formation, through conversion to thiols depending on reaction conditions. Typical cross-linkers may include thiol or amine moieties. Mixed-mode polymerizations are the result of both mechanisms occurring in the same reaction vessel; acrylate and methacrylate groups can undergo mixed mode network formation. Both mechanisms of hydrogel formation can be used to encapsulate live cells, and both mechanisms allow for the reactive incorporation of peptides, proteins and other therapeutics.
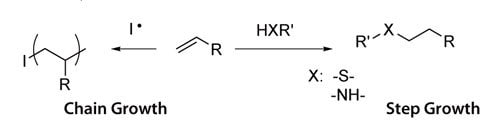
Scheme 1. Chain growth and step growth reactions.
The mesh structure that results from different mechanisms is depicted in Figure 1. In chain growth networks, a kinetic chain is formed at the crosslink site, while in step growth networks, the crosslink sites bear the same functionality as the multifunctional cross-linker, neglecting defects. In both chain and step growth, network defects such as loops, permanent entanglements and dangling chain ends may exist. The chemical identity of the macromer and the mechanism of hydrogel formation are both important as each influences the cross-link density of the hydrogel network. Material properties that are important to 2D and 3D culture are easily controlled through the chemistry of hydrogel formation. As cross-linking density increases, mesh size decreases, swelling ratio decreases, and storage modulus increases. Varying the molecular weight of the PEG macromer results in coarse control over hydrogel properties (large differences in cross-linking density). Varying the reaction mechanism used to produce the hydrogels results in fine control over hydrogel properties (can be used to tune cross-linking density of a system).
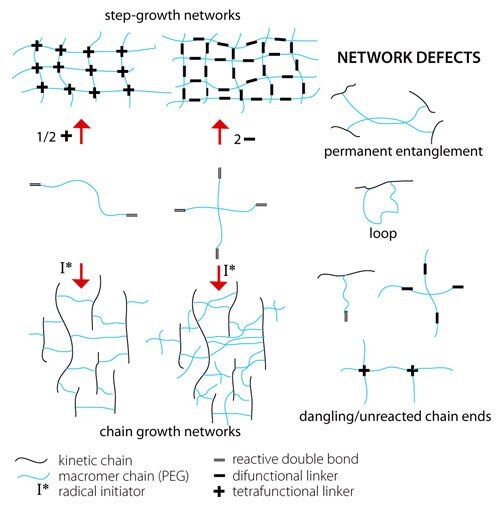
Figure 1. Formation mechanism affects hydrogel network structure and network defects.
In order to use 3D hydrogel scaffolds to study cell differentiation and tissue evolution, it is critical to be able to control the physical and chemical properties of the gel in a spatially and temporally controlled manner.10 Polymeric material properties are typically changed through polymerization/cross-linking (bond forming events) or through controlled degradation and/or release (bond breaking events). Bond forming events typically often use small molecule reagents (initiators, catalysts, monomers, ligands to be conjugated to the material) while bond breaking typically does not rely on exogenous reagents. Small molecules often have more adverse effects in vitro and in vivo than polymeric reagents so many research groups use degradation as a tool for in situ manipulation of polymeric biomaterials.
The mechanism of degradation most commonly utilized in hydrogels is hydrolysis, in which a molecule of water adds to the polymer backbone, causing chain scission. Anhydrides, esters and amides are all susceptible to hydrolysis. Anyhydrides typically hydrolyze too quickly, and the uncatalyzed hydrolysis of amides is too slow, so most hydrogels that degrade hydrolytically utilize ester linkages. In order to obtain hydrolytically degradable hydrogels with physiologically relevant time scales of degradation, researchers typically functionalize PEG with degradable ester linkages using lactide or glycolide segments.
Alcohol chain ends on PEG can initiate ring-opening reactions of 3,6-dimethyl-1,4-dioxane-2,5-dione and 1,4-Dioxane-2,5-dione to generate PEG-lactide and PEG-glycolide, respectively (Scheme 2).11 The ring-opening reaction is typically catalyzed by tin(II)-2-ethylhexanoate,12 although the reaction is also easily accomplished using dimethylaminopyridine as a catalyst,13 which may be easier to remove than the residual tin. The alcohol chain ends of PEG-lactide or PEG-glycolide are easily functionalized with reactive double bonds such as acrylate and methacrylate.
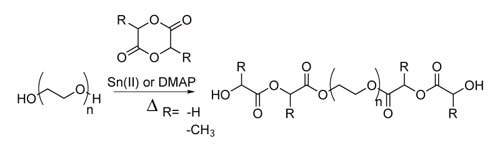
Scheme 2. Synthesis of PEG-lactide and PEG-glycolide.
Although ester linkages are enzymatically degradable, most researchers utilize sequence-specific enzymatic degradation of peptides incorporated into hydrogels rather than non-specific enzymatic degradation of esters and amides. Hubbell′s group pioneered this approach16 by incorporating matrix metalloproteinase (MMP) sensitive linkages into hydrogels via Michael addition of cysteine-functionalized peptides across acrylates, maleimides and vinyl sulfones (Scheme 3).17
MMP-degradable linkages have also been used to tether therapeutic agents into hydrogels. For example, growth factors such as vascular endothelial growth factor (VEG-F) can be released via enzymatic degradation of an MMP-sensitive tether to induce angiogenesis.18
In both hydrolysis and enzymolysis, the rate of degradation is predetermined by the chemistry of the macromer. In hydrolysis, the degradation rate of the material is pre-engineered through the identity (e.g., hydrophobicity or hydrophilicity) and number of the hydrolysable groups, and cannot be changed once the material is fabricated. In enzymolysis, the degradation typically occurs in an area local to the cells producing the enzyme. While hydrolysis and enzymolysis are both effective methods for sustained hydrogel degradation and sustained release of therapeutic agents, the rate of release cannot be adjusted or arrested after the hydrogel is fabricated, and release is not spatially controlled.
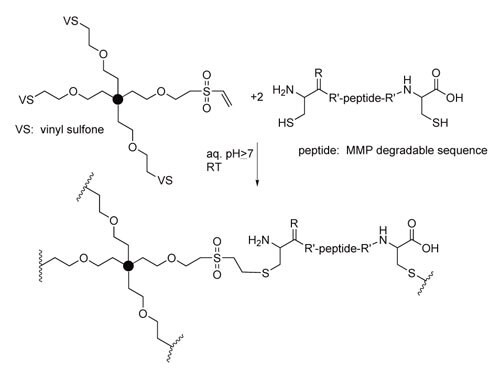
Scheme 3. Enzymatically degradable hydrogels via Michael addition of cysteinecontaining peptides to vinyl sulfone groups.
In contrast to hydrolytically and enzymatically degradable linkages, photodegradable linkages allow precise spatial and temporal control over degradation and release. While many researchers have reported photopolymerizable hydrogels, and photofunctionalizable hydrogels, very few reports exist of biocompatible photodegradable hydrogels. Kloxin and Kasko reported photodegradable hydrogel networks formed from 2-methoxy-5-nitro-4-(1-hydroxyethyl) phenoxybutanoate-containing PEG macromers (Scheme 4)19; the photodegradation behavior of the ortho-nitrobenzyl (o-NB) linker group is well-characterized. Hydrogels formed from the photodegradable macromer show bulk degradation upon exposure to light that is dependent on exposure time, wavelength, and light intensity. When the light is shuttered, degradation is arrested; the sample continues photolyzing once light exposure resumes. hMSCs (human mesenchymal stem cells) encapsulated in a hydrogel containing the photo-releasable cell-adhesive ligand RGDS (Arg-Gly-Asp-Ser) differentiate down the chondrogenic pathway when the RGD is released at day ten (corresponding to the downregulation of fibronectin during chondrogenesis). Surface erosion and through-gel lithography of this degradable hydrogel can be used to form features over a range of lengths scales, from 10-7 m to 10-2 m or larger.20 Partial degradation in a local area results in decreased cross-link density and increased swelling, providing a means to etch softer features onto a hydrogel that protrude out from the gel.
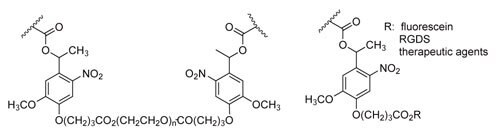
Scheme 4. Photodegradable o-NB moieties incorporated into hydrogel backbone and for therapeutic agent release.
In addition to single photon photolysis, the o-NB containing hydrogels are also susceptible to two-photon photolysis, allowing for 3D etching.19-20 In single photon reactions, any area exposed to the light will react. In contrast, multi-photon lithography should occur only where multiple photons are simultaneously absorbed, which occurs at the focal volume of the light source (inset). Typical wavelengths in single photon lithography of biomaterials range from long wave UV (≥365 nM) into the visible region, while two-photon lithography uses IR light (typically ~740–800 nM). IR light is more biocompatible and less destructive to live tissues and offers greater penetration depth. The probability of twophoton absorption occurring is also tightly limited to the focal point of the focused light, rather than along the entire path of the light, providing 3D control over excitation. Both single- and multi-photon reactions have the potential to pattern materials with features smaller than 500 nM, much smaller than the size of a mammalian cell.21 This represents an unprecedented level of spatial control over hydrogel scaffold structure and chemistry.
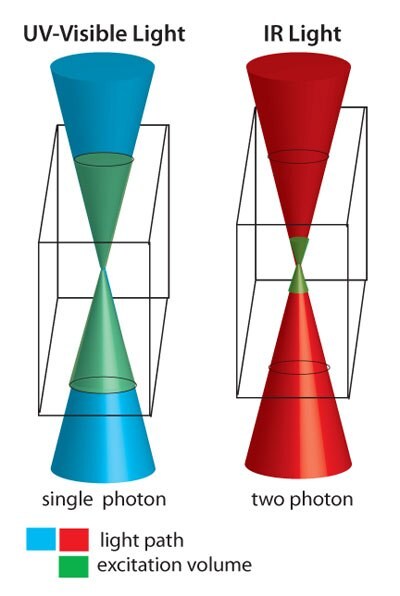
Figure 2.Single photon photolysis (left) occurs in the entire area of the hydrogel exposed to UV-visible light, and two photon photolysis (right) results only in the area where simultaneous absorption of two photons of IR light occurs.
The o-NB linker can also be used to tether therapeutic agents into hydrogels for delivery to live cells. Griffin et al. demonstrated the controlled release of fluorescein tethered into a hydrogel through an o-NB-PEG macromer.22 The release of this model therapeutic as a function of light exposure at multiple wavelengths (365–436 nM), intensities (5–20 mW/cm2) and durations (0–20 minutes) was quantified. While the fastest release occurs at 365 nM (which corresponds to a higher molar absorptivity of the o-NB linker at that wavelength), significant release is also seen at 405 nM; the release is easily modeled from physical constants of the molecules (such as molar absorptivity). Light attenuation allows the facile formation of chemical and mechanical gradients in these systems.
Poly(ethylene glycol) is a readily available, easily modifiable polymer. It has found widespread use in hydrogel fabrication, including as 2D and 3D scaffolds for tissue culture. Degradable linkages are easily introduced into PEG hydrogels. Hydrolytically degradable gels allow for sustained material degradation and/or therapeutic agent release. Degradation and release is cell-dictated in enzymatically degradable gels. Photodegradation allows for real-time user tailored external manipulation of the chemical and physical properties of hydrogels.
To continue reading please sign in or create an account.
Don't Have An Account?