Protein Labeling & Modification
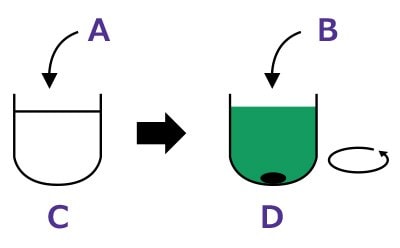
Step 1: Labeling.Dissolve protein in buffer. Add dye and stir for 2 hours.
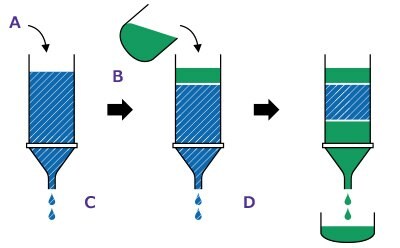
Step 2: Purification.Equilibrate the column. Add reaction mixture, start separation. Collect purified labeled protein solution.
Proteins are composed of polypeptide chains and their biological functions are determined in part by the correct folding, size, and the number of reactive functional groups present throughout the polypeptide chain. The ability to provide site-specific protein modifications provides researchers with the ability to investigate a wide range of properties and their overall biological function. Protein labeling and modifications technologies include adding fluorophores, biotin, and other small molecules to examine protein-protein interactions, protein folding, to investigate the overall protein structure, and their biological function.
Featured Categories
High-quality amino acids, resins, and reagents including Novabiochem® for peptide synthesis, organic chemistry, and custom products.
We offer a comprehensive portfolio of inorganic and metallic nanomaterials, functionalized nanoparticles, and nanomaterial kits for your research needs.
Choose from our comprehensive range of IVD and Biological Staining Commission Certified dry dyes and stains for use with research and clinical specimens.
Discover protein degrader building blocks for creating libraries, like PROTAC® molecules, for efficient target protein degradation in cell screenings.
Fluorescent Protein Labeling
The discovery and wide-spread incorporation of green fluorescent protein (GFP) is a powerful example of this technology. GFP and its counterpart molecules have had an enormous impact on numerous fields of study and our understanding of biological processes. For example, the incorporation of fluorescent tags onto antibodies allows for the detection and quantification of highly specific protein complexes in tissues or the directional immobilization of antibody-protein complexes for ELISA and western blot applications. However, a significant disadvantage of using GFP is that the protein function may be disrupted by the incorporation of additional protein tags of this size. To circumvent this challenge, researchers may use smaller fluorescent tags compared to a GFP, biotin, or incorporate non-natural amino acids that contain biorthogonal functionality.
Enzyme-Protein Conjugation
The incorporation of unique enzymes or probes is another method used by researchers to label proteins. Commonly used enzyme-protein conjugations include alkaline phosphatase (AP), and horseradish peroxidase (HRP). There are numerous advantages for using enzyme-protein labeling technologies as they allow for signal amplification, diverse signal outputs, and there are numerous substrates available for each enzyme. Common signal outputs include fluorescent, chemiluminescent, or colorimetric detection. The variety of these signal outputs make them suitable for immunohistochemistry (IHC) or immunofluorescent (IF)-based detection applications in cells and tissues.
Emerging Protein Labeling Technologies
In the field of disease research and drug discovery, targeted protein degradation technology is being intensely studied by researchers to identify novel drug targets and potential therapeutics. Proteolysis-targeting chimeras technology utilizes bifunctional molecules that are designed on one end to bind to a target disease protein, while the other end binds to an E3 ligase to eliminate the protein from the cell. The combined specificity of these molecules to a wide range of disease targets and their ability to target them for protein degradation using internal cellular protein degradation systems make them a powerful protein labeling technology for disease research.
Visit our document search for data sheets, certificates and technical documentation.
Related Articles
- Learn about the different types of phosphorylation, a vital cellular process that allows for energy storage by converting ADP to ATP. Compare oxidative phosphorylation and substrate level phosphorylation with helpful diagrams.
- Peptidoglycan structure: alternating GlcNAc and N-acetylmuramic acid cross-linked to peptides form the basic backbone.
- Peptide Modifications: N-Terminal, Internal, and C-Terminal
- Chromogenic and fluorogenic derivatives are invaluable tools for biochemistry, having numerous applications in enzymology, protein chemistry, immunology and histochemistry.
- Learn more about Mass Spectrometry or MS including what it is, what it is used for and how it works.
- See All (47)
Related Protocols
- This page how to remove GST tags by enzymatic cleavage using Cytiva products.
- This page shows coupling through the primary amine of a ligand with a NHS-activated Sepharose High Performance, NHS-activated Sepharose 4 Fast Flow and CNBr-activated Sepharose from Cytiva.
- This protocol shows how to remove histidine tags by enzymatic cleavage using Cytiva products.
- This page shows how to purify or remove biotin and biotinylated substances with Streptavidin Sepharose High Performance from Cytiva.
- Introduction to PAGE. Learn about SDS-PAGE background and protocol for the separation of proteins based on size in a poly-acrylamide gel.
- See All (16)
Find More Articles and Protocols
How Can We Help
In case of any questions, please submit a customer support request
or talk to our customer service team:
Email custserv@sial.com
or call +1 (800) 244-1173
Additional Support
- Chromatogram Search
Use the Chromatogram Search to identify unknown compounds in your sample.
- Calculators & Apps
Web Toolbox - science research tools and resources for analytical chemistry, life science, chemical synthesis and materials science.
- Customer Support Request
Customer support including help with orders, products, accounts, and website technical issues.
- FAQ
Explore our Frequently Asked Questions for answers to commonly asked questions about our products and services.
To continue reading please sign in or create an account.
Don't Have An Account?



