Removal of GST Tag by Enzymatic Cleavage
Removal of the GST tag is often necessary to be able to perform functional or structural studies of the target protein. Tagged proteins containing a PreScission Protease, thrombin, or Factor Xa recognition site can be cleaved either while bound to Glutathione Sepharose® or in solution after elution. Cleavage releases the target protein from the column and allows elution using the binding buffer. The GST moiety remains bound to the medium.
PreScission Protease itself has a GST tag and therefore will bind to Glutathione Sepharose; it will thus not co-elute and contaminate the cleaved target protein. Cleavage with PreScission Protease is very specific, and maximum cleavage is obtained in the cold (the protein is most active at 4 °C), thus improving the stability of the target protein.
If thrombin or Factor Xa are used for cleavage of the tag, a convenient way to remove these enzymes is to connect in series one GSTrap™ FF column and one HiTrap™ Benzamidine FF (high sub) column. During the elution the cleaved product passes directly from the GSTrap™ into the HiTrap™ Benzamidine FF (high sub). The cleaved target protein passes through the HiTrap™ Benzamidine FF (high sub) column but the proteases bind. Thus in a single step the enzymes are removed and a pure cleaved target protein is achieved (Figure 5.17). Note, however, that thrombin and Factor Xa may produce a less specific cleavage than PreScission Protease and that sometimes the target protein can be fragmented itself.
1 PreScission Protease is a tagged protein of glutathione S-transferase and human rhinovirus type 14 3C protease.
The amount of enzyme, temperature, and length of incubation required for complete digestion varies according to the specific GST-tagged protein produced. Optimal conditions should always be determined in pilot experiments.
If protease inhibitors (Table 5.6) have been used in the lysis solution, they must be removed prior to cleavage with PreScission Protease, thrombin, or Factor Xa. (The inhibitors will usually be eluted in the flowthrough when sample is loaded onto a GSTrap™ column.)
Cleavage of tagged proteins is most commonly performed on milligram quantities of tagged protein suitable for purification on GSTrap™ columns. Protocols that follow describe manual cleavage and purification using a syringe and a 1 mL or 5 mL GSTrap™ column. The protocols can be adapted for use with GST MultiTrap™ or GST SpinTrap™ columns to work at smaller scales.
For quick scale-up of purifications, two or three GSTrap™ columns can be connected in series (back pressure will be higher). Further scaling-up is possible using GSTPrep™ FF 16/10 columns or columns packed by the user. Protocols below are included for column or batch format using Glutathione Sepharose® 4 Fast Flow, but this medium can easily be replaced with Glutathione Sepharose® High Performance or Glutathione Sepharose® 4B depending on what is the preferred chromatography medium in the lab.
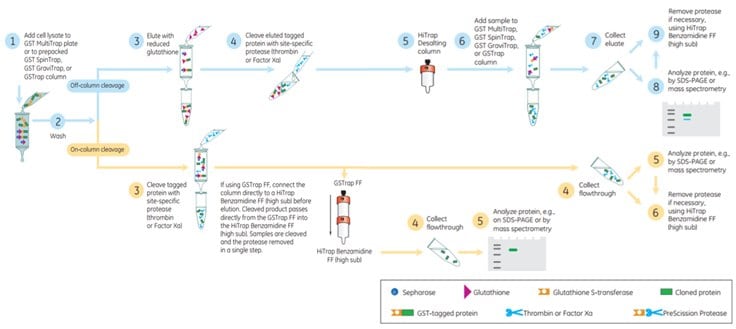
Figure 5.17Cleavage of GST Tag using Thrombin or Factor Xa.
Purification and Cleavage
The protocol below is an example optimized for 8 mg of target protein. It is worth estimating how much target protein is applied to the column, as this allows one to minimize the amount of protease added.
- Fill the syringe or pump tubing with distilled water. Remove the stopper and connect the column to the syringe (use the connector supplied), laboratory pump, or chromatography system “drop to drop” to avoid introducing air into the system.
- Remove the snap-off end at the column outlet.
- Wash out the ethanol with 3 to 5 column volumes of distilled water.
- Equilibrate the column with at least 5 column volumes of binding buffer. Recommended flow rates are 1 mL/min (1 mL column) and 5 mL/min (5 mL column).
- Apply the pretreated sample using a syringe fitted to the Luer connector or by pumping it onto the column. For best results, use a flow rate of 0.2 to 1 mL/min (1 mL column) and 0.5 to 5 mL/min (5 mL column) during sample application.
- Wash with binding buffer (generally at least 5 to 10 column volumes) until the absorbance reaches a steady baseline or no material remains in the effluent. Maintain a flow rate of 1 to 2 mL/min (1 mL column) and 5 to 10 mL/min (5 mL column) for washing.
- For PreScission Protease and Factor Xa, wash the column with 10 column volumes of cleavage buffer.
For thrombin, proceed to step 8b.
For Factor Xa, proceed to step 8c. - a) Prepare the PreScission Protease mix:
For GSTrap™ FF 1 mL columns, mix 80 µl (160 units) of PreScission Protease with 920 µl of PreScission cleavage buffer at 5 °C.
For GSTrap™ FF 5 mL columns, mix 400 µl (800 units) of PreScission Protease with 4.6 mL of PreScission cleavage buffer at 5 °C.
b) Prepare the thrombin mix:
For GSTrap™ FF 1 mL columns, mix 80 µl (80 units) of thrombin solution with 920 µl of PBS.
For GSTrap™ FF 5 mL columns, mix 400 µl (400 units) of thrombin solution with 4.6 mL of PBS.
c) Prepare the Factor Xa mix:
For GSTrap™ FF 1 mL columns, mix 80 µl (80 units) of Factor Xa solution with 920 µl of Factor Xa cleavage buffer.
For GSTrap™ FF 5 mL columns, mix 400 µl (400 units) of Factor Xa solution with 4.6 mL of Factor Xa cleavage buffer. - Load the protease mix onto the column using a syringe and the connector supplied. Seal the column with the top cap and the stopper supplied.
- For PreScission Protease, incubate the column at 5 °C for 4 h.
For thrombin and Factor Xa, incubate the column at room temperature (22 °C to 25 °C) for 2 to 16 h.
The incubation times are starting points and may need to be changed for an optimal yield of cleaved target protein.
- Fill a syringe with 3 mL (1 mL column) or 15 mL (5 mL column) of cleavage buffer. Remove the top cap and stopper from the column and attach the syringe. Avoid introducing air into the column.
- Begin elution of the cleaved target protein. Maintain flow rates of 1 to 2 mL/min (1 mL column) or (5 mL column), and collect the eluate (0.5 to 1 mL/tube for 1 mL column, 1 to 2 mL/tube for 5 mL column).
For PreScission Protease: The eluate will contain the protein of interest, while the GST moiety of the tagged protein and the PreScission Protease (also GST-tagged) will remain bound to the GSTrap™ column. This means that the protein of interest will not be contaminated with protease and thus no additional purification will be required to purify the target protein from the protease.
For thrombin and Factor Xa: The eluate will contain the protein of interest and thrombin or Factor Xa, respectively, while the GST moiety of the tagged protein will remain bound to the GSTrap™ column. Thrombin or Factor Xa can be removed from the protein of interest in one step using a HiTrap™ Benzamidine FF (high sub) column in series after the GSTrap™ column. In this process, the cleaved, tagged protein and thrombin or Factor Xa is washed from the GSTrap™ column onto the HiTrap™ Benzamidine FF (high sub) column. This second column captures the thrombin or Factor Xa, thus enabling the collection of free protein in the eluent. Refer to Figure 5.20 for an example of the purification and on-column cleavage of GST-tagged SH2 domain using thrombin and GSTrap™ FF, with sample cleanup accomplished using HiTrap™ Benzamidine FF (high sub) column in series with GSTrap™ FF. Removal of thrombin and Factor Xa using HiTrap™ Benzamidine FF (high sub) later on in this chapter for the procedure.
Appendix 2 (Characteristics of Glutathione Sepharose® products) for details on regenerating the GSTrap™ column for subsequent purifications.
Application Examples
1. Purification of human hippocalcin using GSTrap™ FF columns in series with on-column cleavage by PreScission Protease
The gene for human hippocalcin, a member of the neuron-specific calcium-binding protein family, was cloned into a pGEX vector containing a PreScission Protease site adjacent to the GST tag. The expressed tagged protein was captured on a GSTrap™ FF 1 mL column. The column was then incubated overnight at 4 °C and for an additional 2 h at room temperature with PreScission Protease (which is GST-tagged itself). Following on-column cleavage, a second GSTrap™ FF 1 mL column was placed in series after the first to remove any PreScission Protease, uncleaved GST-tagged protein, or free GST tag that could co-elute with the sample during the additional wash with binding buffer (Figure 5.18). For every gram of wet E. coli cells, 10 mg of pure, untagged hippocalcin was obtained.
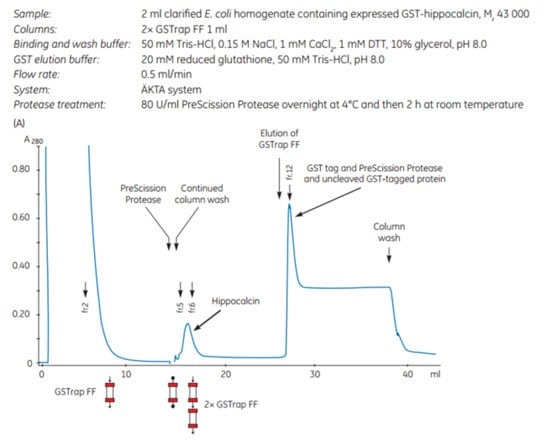

Figure 5.18.Purification of human hippocalcin-GST-tagged protein with on-column cleavage and post-cleavage removal of PreScission Protease using GSTrap™ FF columns. (A) Chromatogram showing purification of hippocalcin. (B) SDS-PAGE analysis of various sample processing steps. ExcelGel SDS Gradient, 8–18, Coomassie blue staining.
2. Automatic removal of the GST tag with PreScission Protease
This example of automated tag removal uses ÄKTAxpress. All multistep purification protocols in ÄKTAxpress can be combined with automated on-column tag cleavage. Tag cleavage is always performed on the affinity column prior to further purification steps. When the cleaved protein has been eluted, the affinity column is regenerated and affinity tag, tagged protease, and remaining uncleaved protein are collected in a separate outlet. The procedure involves binding the tagged protein, injection of protease, incubation, elution of cleaved protein, and collection in capillary loop(s), followed by further purification steps.
The example in Figure 5.19 shows purification results for a GST-tagged protein, GST-purα (Mr 61 600), expressed in E. coli. The Mr of the cleaved product is 35 200. After harvest, cell lysis was performed by sonication. The samples were clarified by centrifugation prior to sample loading.
AC and SEC were performed on ÄKTAxpress using columns as indicated in the figure. The purity of each sample was analyzed by SDS-PAGE (Coomassie staining). The reduced samples were applied on an ExcelGel SDS-polyacrylamide gel.
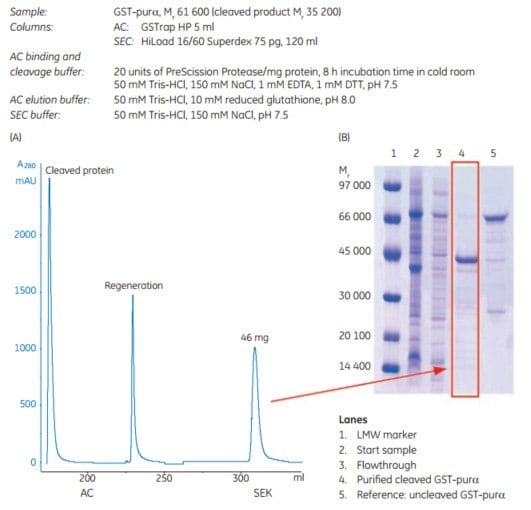
Figure 5.19.(A) Two-step protocol for automatic GST-tagged protein cleavage with PreScission Protease and purification. (B) Analysis by SDS-polyacrylamide gel (Coomassie staining) of the untagged target protein after purification and cleavage.
3. Purification and on-column cleavage of GST-tagged SH2 domain using thrombin and GSTrap™ FF. Direct removal of thrombin using HiTrap™ Benzamidine FF (high sub) column in series with GSTrap™ FF
The following application describes the purification of GST-SH2 (Mr 37 000) on a GSTrap™ FF 1 mL column, followed by on-column cleavage with thrombin (Figure 5.20). After the thrombin incubation step, a HiTrap™ Benzamidine FF (high sub) 1 mL column was placed in series after the GSTrap™ FF column. As the columns were washed with binding buffer and later with high-salt buffer, the cleaved SH2-tagged protein and thrombin were washed from the GSTrap™ FF column onto the HiTrap™ Benzamidine FF (high sub) column. Thrombin was captured by this second column, thus enabling the collection of pure thrombin-free untagged target protein in the eluent (Figure 5.20A). Complete removal of thrombin was verified using the chromogenic substrate S-2238 (Chromogenix, Haemochrom Diagnostica AB; supplier in US is DiaPharma) for detection of thrombin activity (Fig 5.20B). This entire procedure could be completed in less than one day.
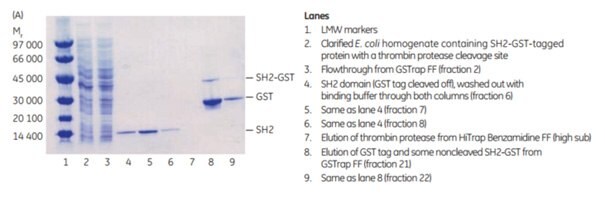
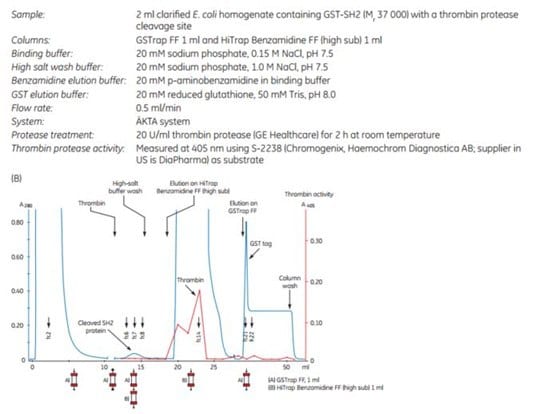
Figure 5.20.Purification of GST-SH2 GST-tagged protein with on-column cleavage and post-cleavage removal of thrombin using GSTrap™ FF and HiTrap™ Benzamidine FF (high sub) columns. (A) SDS-PAGE analysis of various sample processing steps. ExcelGel SDS Gradient 8–18, Coomassie blue staining. (B) Chromatogram (blue: absorbance at 280 nm) and thrombin activity curve (red) demonstrating all steps in the purification of the SH2 domain.
4. On-column cleavage of a GST-tagged protein using thrombin on a GSTrap™ FF column
To demonstrate the efficiency of on-column cleavage in conjunction with purification, a GST-tagged protein containing the recognition sequence for thrombin, was applied to GSTrap™ FF 1 mL. After washing, the column was filled by syringe with 1 mL of thrombin solution (20 U/mL in PBS, pH 7.3) and sealed using the supplied connectors. After incubation for 16 h at room temperature, the target protein minus the GST moiety was eluted using PBS, pH 7.3, and the bound GST was subsequently eluted using elution buffer (Figure 5.21). The cleavage reaction yield was 100%. Intact GST-tagged protein was not detected in the eluate by SDS-PAGE and silver staining (Figure 5.21C, lane 5).
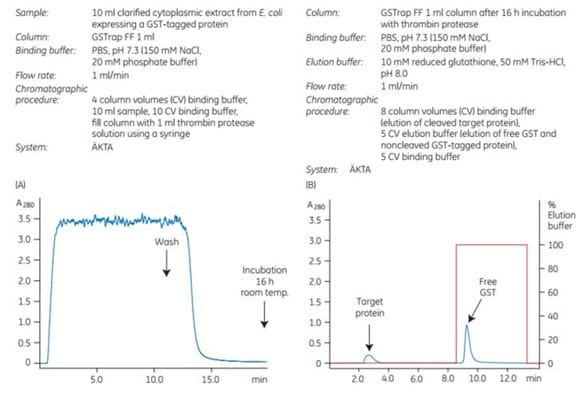
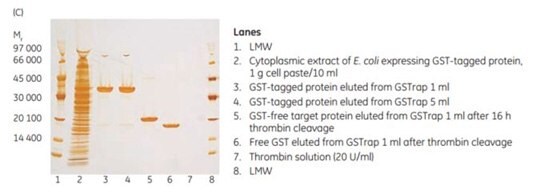
Figure 5.21.On-column thrombin cleavage of a GST-tagged protein. (A) Equilibration, sample application, and washing of a GST-tagged protein on GSTrap™ FF 1 mL were performed using ÄKTA chromatography system. After washing, the column was filled by syringe with 1 mL of thrombin (20 U/mL) and incubated for 16 h at room temperature. (B) GST-free target protein was eluted using PBS, pH 7.3. GST was eluted using 10 mM reduced glutathione. (C) SDS-PAGE followed by silver staining. The GST-free target protein fraction also contained a small amount of thrombin not detectable by SDS-PAGE (lane 6). The thrombin can be removed using a HiTrap™ Benzamidine FF (high sub) column.
Purification and Cleavage
The protocol below is an example optimized for 8 mg of target protein. It is worth estimating how much target protein is applied to the column, as this allows one to minimize the amount of protease added.
- Fill the syringe or pump tubing with distilled water. Remove the stopper and connect the column to the syringe (use the connector supplied), laboratory pump, or chromatography system “drop to drop” to avoid introducing air into the system.
- Remove the snap-off end at the column outlet.
- Wash out the ethanol with 3 to 5 column volumes of distilled water.
- Equilibrate the column with at least 5 column volumes of binding buffer. Recommended flow rates are 1 mL/min (1 mL column) and 5 mL/min (5 mL column).
- Apply the pretreated sample using a syringe fitted to the Luer connector or by pumping it onto the column. For best results, use a flow rate of 0.2 to 1 mL/min (1 mL column) and 0.5 to 5 mL/min (5 mL column) during sample application.
- Wash with binding buffer (generally at least 5 to 10 column volumes) until the absorbance reaches a steady baseline or no material remains in the effluent. Maintain a flow rate of 1 to 2 mL/min (1 mL column) and 5 to 10 mL/min (5 mL column) for washing.
- Elute the GST-tagged protein with 5 to 10 column volumes of elution buffer. Maintain flow rates of 1 to 2 mL/min (1 mL column) or 1 to 5 mL/min (5 mL column). Collect the eluate (0.5 to 1 mL/tube for 1 mL column, 1 to 2 mL/tube for 5 mL column). Pool fractions containing the GST-tagged protein (monitored by UV absorption at A280).
- Remove the free reduced glutathione from the eluate using a quick buffer exchange on a desalting column (Chapter 11), depending on the sample volume.
- a) For PreScission Protease, add 1 µl (2 units) of PreScission Protease for each 100 µg of tagged protein in the buffer-exchanged eluate.
b) For thrombin and Factor Xa, add 10 µl (10 units) of thrombin or Factor Xa solution for each mg of tagged protein in the buffer-exchanged eluate. - a) For PreScission Protease, incubate at 5 °C for 4 h.
b) For thrombin and Factor Xa, incubate
The incubation times are starting points and may need to be changed for an optimal yield of cleaved target protein.
- Once digestion is complete, apply the sample to an equilibrated GSTrap FF column as described above (steps 1 to 6) to remove the GST moiety of the tagged protein.
For PreScission Protease: The flowthrough will contain the protein of interest, while the GST moiety of the tagged protein and the PreScission Protease will remain bound to the GSTrap column. This means that the protein of interest will not be contaminated with protease and thus no additional purification will be required to purify the target protein from the protease.
For thrombin and Factor Xa: The flowthrough will contain the protein of interest and thrombin or Factor Xa, respectively, while the GST moiety of the tagged protein will remain bound to the GSTrap column. The thrombin or Factor Xa can be removed from the protein of interest in one step using a HiTrap Benzamidine FF (high sub) column in series after the GSTrap column. In this process, the cleaved, tagged protein and thrombin or Factor Xa is washed from the GSTrap column onto the HiTrap Benzamidine FF (high sub) column. This second column captures the thrombin or Factor Xa, thus enabling the collection of pure protease-free protein in the eluent. Removal of thrombin and Factor Xa using HiTrap Benzamidine FF (high sub) for the procedure.
Appendix 2 (Characteristics of Glutathione Sepharose products) for details on regenerating the GSTrap column for subsequent purifications.
Cleavage and purification of GST-tagged protein bound to Glutathione Sepharose in batch mode
Glutathione Sepharose High Performance, Glutathione Sepharose 4 Fast Flow, and Glutathione Sepharose 4B can all be used for cleavage and purification of GST-tagged proteins in batch.
Preparation of Glutathione Sepharose Chromatography Media and Binding of Protein
Glutathione Sepharose media are supplied in 20% ethanol. The media are used at a final slurry concentration of 50%.
- Determine the bed volume of Glutathione Sepharose required for your purification.
- Gently shake the bottle to resuspend the slurry.
- Use a pipette or measuring cylinder to remove sufficient slurry for use and transfer to an appropriate container/tube.
- Sediment the chromatography medium by centrifugation at 500 × g for 5 min. Carefully decant the supernatant.
- Wash the Glutathione Sepharose by adding 5 mL of PBS per 1 mL of 50% slurry.
Glutathione Sepharose must be thoroughly washed with PBS to remove the ethanol storage solution because residual ethanol may interfere with subsequent procedures.
- Sediment the chromatography medium by centrifugation at 500 × g for 5 min. Carefully decant the supernatant.
- Repeat steps 5 and 6 once for a total of two washes.
- Add the cell lysate to the prepared Glutathione Sepharose and incubate for at least 30 min at room temperature, using gentle agitation such as end-over-end rotation.
Purification and Cleavage
Assume 8 mg of GST-tagged protein bound per mL of chromatography medium.
- Wash the tagged-protein-bound Glutathione Sepharose with 10 bed volumes of cleavage buffer. Bed volume is equal to 0.5× the volume of the 50% Glutathione Sepharose slurry used.
- a) Prepare the PreScission Protease mix:
For each mL of Glutathione Sepharose bed volume, prepare a mixture of 80 µl (160 units) of PreScission Protease and 920 µl of cleavage buffer at 5 °C.
b) Prepare the thrombin mix:
For each mL of Glutathione Sepharose bed volume, prepare a mixture of 80 µl (80 units) of thrombin and 920 µl of cleavage buffer.
c) Prepare the Factor Xa mix:
For each mL of Glutathione Sepharose bed volume, prepare a mixture of 80 µl (80 units) of Factor Xa and 920 µl of cleavage buffer. - Add the mixture to the Glutathione Sepharose. Gently shake or rotate the suspension end-over-end.
- a) For PreScission Protease, incubate at 5 °C for 4 h.
b) For thrombin or Factor Xa, incubate at room temperature (22 °C to 25 °C) for 2 to 16 h.
The incubation times in steps 4a and 4b are starting points and may need to be changed for an optimal yield of cleaved target protein.
- Following incubation, wash out the untagged protein with approximately three bed volumes of cleavage buffer. Centrifuge the suspension at 500 × g for 5 min to pellet the Glutathione Sepharose. Carefully transfer the eluate to a tube.
For PreScission Protease: The eluate will contain the protein of interest, while the GST moiety of the tagged protein and the PreScission Protease will remain bound to the Glutathione Sepharose. This means that the protein of interest will not be contaminated with protease and thus no additional purification will be required to purify the target protein from the protease.
For thrombin and Factor Xa: The eluate will contain the protein of interest and thrombin or Factor Xa, respectively, while the GST moiety of the tagged protein will remain bound to the Glutathione Sepharose. The thrombin or Factor Xa can be removed from the protein of interest using HiTrap Benzamidine FF (high sub). This column captures the thrombin or Factor Xa, thus enabling the collection of pure protease-free protein in the eluent. procedure below.
Removal of thrombin and Factor Xa using HiTrap Benzamidine FF (high sub)
Reagents required
Binding buffer:
0.05 M Tris-HCl, 0.5 M NaCl, pH 7.4
Elution buffer alternatives for eluting the protease:
0.05 M glycine-HCl, pH 3.0
10 mM HCl, 0.5 M NaCl, pH 2.0
20 mM p-aminobenzamidine in binding buffer (competitive elution) 8 M urea or 6 M Gua-HCl (denaturing solutions)
Recommended flow rates are 1 mL/min (1 mL column) or 5 mL/min (5 mL column).
- Fill the syringe or pump tubing with distilled water. Remove the stopper and connect the column to the syringe (use the connector supplied), laboratory pump, or chromatographic system “drop to drop” to avoid introducing air into the column.
- Remove the snap-off end.
- Wash the column with 5 column volumes of distilled water to remove the storage buffer (0.05 M acetate buffer, pH 4, containing 20% ethanol).
- Equilibrate the column with 5 column volumes of binding buffer.
- Apply the sample using a syringe fitted to the Luer connector or by pumping it onto the column. Recommended flow rates for sample application are 1 mL/min for 1 mL column and 5 mL/min for 5 mL column. Collect the flowthrough and reserve. It contains the protease-depleted material to be saved. Apply a small volume of extra binding buffer to collect all desired material from the column.
- Wash the column with 5 to 10 column volumes of binding buffer, collecting fractions (0.5 to 1 mL fractions for 1 mL column and 1 to 3 mL fractions for 5 mL column) until no material appears in the effluent (monitored by UV absorption at 280 nm).
- Pool fractions from flowthrough and/or wash that contain the thrombin- or Factor Xa-free material (monitored by UV absorption at 280 nm).
- For reuse of column, elute the bound protease with 5 to 10 column volumes of the elution buffer of choice. If the eluted thrombin or Factor Xa is to be retained for reuse, buffer exchange the fractions containing the protease using a desalting column (Chapter 11, Desalting/buffer exchange and concentration).
- If a low pH elution buffer has been used, collect fractions in neutralization buffer.
- After all protease has been eluted, wash the column with binding buffer so it is ready for reuse.
Thrombin activity can be followed by taking aliquots of the fractions and measuring at 405 nm using S-2238 (Chromogenix, Haemochrom Diagnostica AB; supplier in US is DiaPharma) as substrate.
Materials
To continue reading please sign in or create an account.
Don't Have An Account?