Validation of Shorter Protocol for Detection of Salmonella enterica subsp. enterica in Peanut Butter Samples Followed by a rRNA Detection System
Abstract
Food-borne pathogens Salmonella is commonly evaluated in manufacturing of peanut butter and other food products. For the HybriScan®D Salmonella Test (Product No. 55662) the ISO based enrichment method is recommended. That means sample pre-enrichment for 18 hours at 37 °C in buffered peptone water (BPW) followed by a selective enrichment step in Rappaport-Vassiliadis (RV) Broth for 24 hours at 41 °C. HybriScan®D Salmonella is a rRNA sandwich hybridisation detection system which needs at least 500 cfu/ml for the assay. This rapid molecular test system is desirable for the detection of Salmonella species like S. Enteritidis, S. Typhimurium, S. Typhi and S. Paratyphi. Results of this study on a rapid test kit demonstrate that Salmonella enterica subsp. enterica (ATTC 13311) can be detected and identified in a shorter time even in a difficult sample matrix like peanut butter.
Introduction
Peanut butter consist of about 20% carbohydrates, 25% proteins and 50% fat, Salmonella cells are just a very small component of the overall sample material and may be attached within the food matrix as single cells or clumps of cells. Normally before rapid detection methods can be used successfully, it is usually necessary to separate the target cells from the food matrix and from the background microflora. But even the HybriScan is as well a rapid molecular biological system it is practically insensitive to the sample matrix.
HybriScan®D Salmonella test is based on the detection of target molecules from the microorganisms of interest by means of specific capture and detection probes in a so-called sandwich hybridization. The hybridization reaction of the target molecules with the Biotin-labeled capture and a DIG-labeled detection probe takes place in a streptavidin coated microtiter plate (Figure 1).
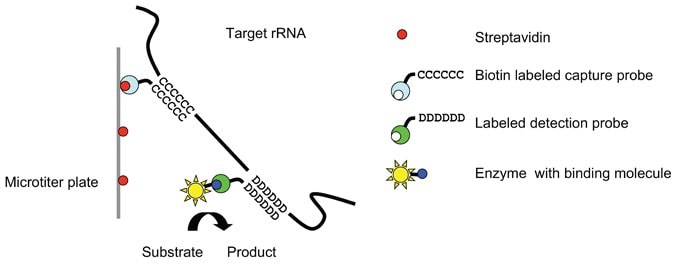
Figure 1.Sandwich Hybridization between Target rRNA Capture and Detection Probe.
After coupling of the target molecule to the microtiter plate, an enzyme is attached in a subsequent incubation step.
After several washing steps, reaction with a color substrate gives blue coloration that changes into yellow color after the
addition of a stop solution. The yellow color enables highly sensitive photometric measurement at 450 nm (Figure 2).
Comparison is made with the standard solutions contained in the test kit.
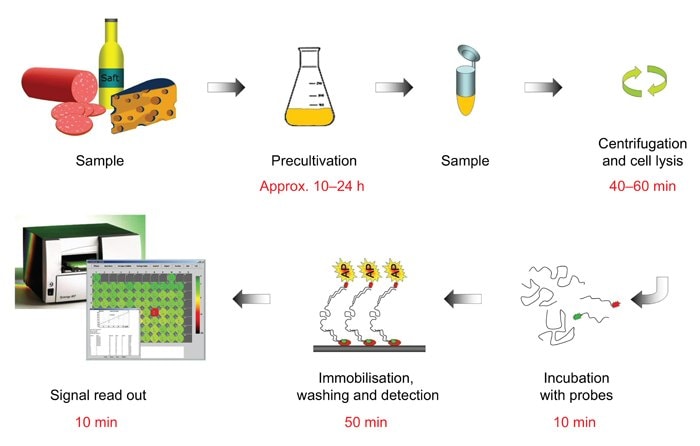
Figure 2.Work flow – HybriScan®D Salmonella.
Experimental
The matrix to be examined was Peanut Butter (9 different brand codes with 12 different code dates). The pre-enrichment time described in the protocol of the HybriScan®D Salmonella test takes 42 hours. In this experiment one target was to reduce the enrichment time to 24 hours in total. The cultivation time of the pre-enrichment peptone water culture took 18 hours; the incubation time of the selective enrichment culture in Rappaport-Vassiliades Enrichment Broth was shortened to 6 hours. Per peanut butter batch, one sample wasn't inoculated and carried as a negative control (N.C.),9 to 10 samples were inoculated with Salmonella enterica subsp. enterica (ATTC 13311). Each Peanut Butter sample was treated as follows:
- 25 g of Peanut Butter (except the negative controls) were inoculated with 1 to 5 cells of Salmonella enterica subsp. enterica.
- 225 mL buffered peptone water were added to each sample, the mixture was homogenised for 1 minute in a Stomacher and the sample were incubated for 18 hours at 37 °C.
- After 18 hours of incubation 0.1 mL of the pre-enrichment peptone water culture were transferred to 10 mL
Rappaport-Vassiliades Enrichment Broth. The selective main enrichment was conducted for 6 hours at 41 °C.
Cell lysis and the HybriScan®D Salmonella assay were completed as described in the HybriScan®D Salmonella test protocol. In addition, each negative control and inoculated sample were tested for Salmonella according to EN ISO 6579:2002.
Results
Evaluation of the samples was performed using the following formula as described in the HybriScanD® Salmonella assay:
Sample O.D.% = (O.D.sample ̶ O.D.N.C.) ̸ (O.D.P.C. ̶ O.D.N.C.) × 72.1%
P.C. positive control (S3)
N.C. negative control (S1)
Samples with O.D.% values under 10 are considered negative.
Samples with O.D.% values from 10 to <20 are considered questionable.
Samples with O.D.% values ≥10 are considered positive.
Of the 107 inoculated with Salmonella enterica peanut butter samples, 106 were identified as clearly contaminated with Salmonella by the use of HybriScan®D Salmonella assay. The result of 1 sample was considered questionable. All negative controls gave negative results in the HybriScan®D Salmonella assay. Table 1– 4.
S1= Standard1 0 cells/10 µL
S2=Standard2 10.000 cells/µL
S3=Standard3 30.000 cells/10µL
S4= Standard4 90.000 cells/10µL
N.C.= Negative control: sample was not inoculated with Salmonella spp.
Materials
To continue reading please sign in or create an account.
Don't Have An Account?