Antibody Enhanced-Validation Strategies
Additional Validation Data to Ensure Antibody Reproducibility, Specificity and Performance
Our Enhanced Antibody Validation Efforts
Enhanced antibody validation is an assay or method that provides researchers with additional assurance that the antibody specificity for the target antigen agrees with previously defined expression data. Many concerned scientists have called for more rigorous evidence that demonstrates commercial antibodies specifically bind to the intended target. To answer the calls for more stringent corroboration of immunodetection application-based antibody validation, we are pleased to offer enhanced validation for many antibodies in our portfolio. Enhanced validation (EV) data that is now represented on our product pages are available for those antibodies in our catalog that carry the ‘EV’ symbol. All antibodies we offer that carry the ‘EV’ symbol will be accompanied by data resulting from one or more of these enhanced validation strategies.
Current commercial antibody validation practices
It is common for commercial antibody developers to assess the on-target binding of an antibody during post immunization screening solely by Western blot (WB) or immunohistochemistry (IHC). This screen ensures that the antibody in development is, at this initial stage, recognizing the expected target and performing similarly to previous lots, if applicable. However, this simple screening is not adequate to assure application suitability and true lot-to-lot consistency.
We therefore subsequently test in as many additional immunodetection applications as practical in samples chosen to be relevant to the intended use of the product. These include immunohistochemistry, immunocytochemistry (ICC), Western blot, ELISA, immunoprecipitation, and more. This in-depth application testing can help assess the antibody’s specificity for the target and provides contextually relevant validation in applications and samples most likely to be used by our customers. This application-specific data should be reviewed by the researcher and appropriately assessed for the researchers intended use. Beyond review of the vendor generated application data, critical review of the epitope, species reactivity, clonality, appropriate host species, and development of appropriate controls are critical responsibilities of the researcher in the selection and use of antibody product.
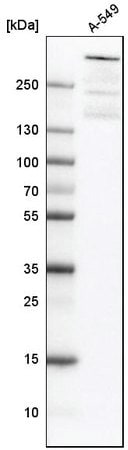
Western blot analysis using anti-ATRX antibody in human cell line A-549. Cat. No. HPA001906
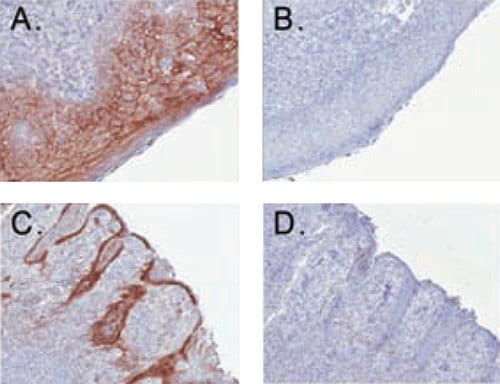
Enhanced Antibody Validation for Immunohistochemistry using anti-desmoglein in human tonsil tissue. Membrane junction immunoreactivity was observed in the stratified squamous epithelium (A) as well as the epithelial cells lining the crypts of the human tonsil (C). Treatment of the same tissues with negative control reagent (B, D) resulted in no detectable signal.
Cat. No. MABT118
Recombinantly Expressed Antibodies
Enhanced validation with recombinant monoclonal antibodies
Characterization provided by the supplier of antibody binding in relevant research applications, followed by appropriate selection and optimization by the researcher is essential for confidence in antibody performance and the scientific conclusions that result from their use. To validate an antibody, it must be shown to be specific, reproducible, and have high-affinity to the target protein.
Recombinant antibodies specificity
Non-specific antibodies may recognize additional proteins to the one they are designed to detect and often bind to more than one target. Our recombinant antibodies represent an entirely new generation of monoclonal antibody. Manufactured using a recombinant expression system, our recombinant antibodies are defined down to the level of the DNA sequence that encodes them, manufactured in engineered 'recombinant' cells and purified for exquisite specificity.
Recombinant antibodies reproducibility and high-affinity
With potential batch-to-batch variability for antibodies traditionally produced in animals, the life sciences are currently challenged by reproducibility. Challenges for antibodies produced in living organisms include loss of expression, altered heavy and light chain expression in the host, mutation, frequent testing or even re-cloning. Recombinant antibodies, defined at the DNA sequence that produces them, result in robust and highly reproducible lot-to-lot consistency which circumvents much of the variability that is introduced by production in animals. As such, recombinant antibodies are consistently available with the same optimal performance and will never suffer from supply issues or clonal drift that antibodies produced in animals often do. Antibodies that bind to the target protein may recognize different epitopes (linear or conformational) or different isoforms for the same target. Our recombinant antibodies provide robust, high-affinity antibody performance for each assay that the antibody has been validated for use.
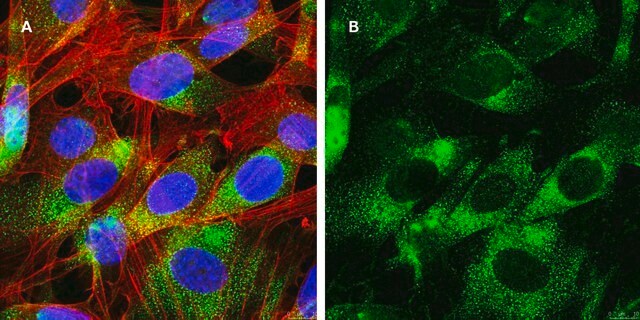
Recombinant antibodies offer similar performance as traditional polyclonal and monoclonal antibodies with the enhanced specificity and reproducibility through recombinant technology and expression. (A) Immunofluorescent analysis of NIH/3T3 cell lines was performed using a 1:100 dilution of Cat. No. ZRB04338, Anti-EGFR, clone 11E19 ZooMAb® Rabbit Monoclonal and visualized with a Goat Anti-Rabbit secondary antibody conjugated to Alexa Fluor® 488 (Green). Actin filaments have been labeled with phalloidin (Red). Nucleus is stained with DAPI (Blue). This antibody positively stains the plasma membrane, cytoplasm.
Genetic Strategies
Genetic Strategies – Demonstrating antibody specificity through knockout/knockdown methods
Expected Results: Diminished or absence of band in Western blotting in knockdown/knockout validation.
Our genetic strategies validation attempts to ensure specificity of protein detection by eliminating or reducing the expression of the target protein through gene editing or gene interference methods and comparing the antibody recognition for such a sample compared to the wild type. The specificity of the antibody can subsequently be assessed through WB or ICC staining of the cell sample that has been altered to reduce or eliminate the target protein expression. The altered protein expression is then compared to the wild type sample that has been demonstrated to express the target protein.
CRISPR/Cas9 genome editing technology enables the creation of cell lines with a specific gene or genes permanently excised so that expression of the corresponding protein is ‘knocked out’. For antibody validation this poses a potentially powerful tool. In simplest terms, a sample with the target “knocked out” should show no antibody staining as the intended target has been genetically removed and the corresponding wild type sample should demonstrate recognition of the target. Degrees of non-specificity can be assessed by comparing these two sample types.
Alternatively, RNA interference (RNAi) can be used to “knock down” or suppress a specific gene to obtain a reduced protein expression. As with the knockout approach, one would compare the genetically altered “knockdown” sample to the wild type. In contrast with genetic knockout, antibody reactivity signal observed in RNAi-altered samples should be reduced in intensity compared to the wild type and often not completely abolished.

Example of genetic validation by RNAi knockdown. Western blot analysis using an anti-ATRX antibody in A-549 cells transfected with control siRNA and two target-specific siRNA probes. Downregulation of ATRX in siRNA samples confirms specificity of antibody.
Independent Antibody Verification
Independent Antibody Verification – Demonstrating antibody specificity through the use of multiple antibodies against target in IHC or ICC.
Expected results: All antibodies should show similar staining patterns or experimental results.
The validity of results obtained with an antibody in a given immunoapplication may be supported by showing that the same results are obtained using the identical protocol with a different antibody raised against the same target. At least two antibodies with non-overlapping epitopes are applied across a panel of samples, such as sections from the same tissue. This approach has the added advantage of enabling validation of both antibodies used for comparison of binding characteristics.
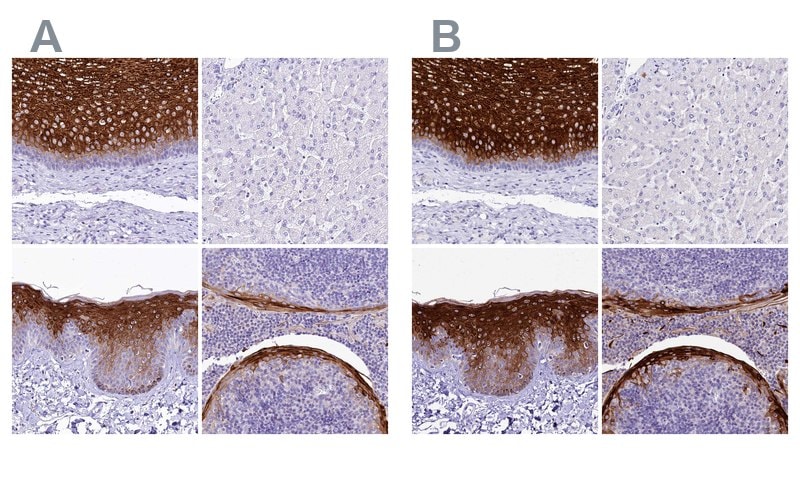
Immunohistochemical staining of human esophagus, liver, skin, and tonsil using Anti-A2ML1 antibody HPA038847 (A) shows similar protein distribution across tissue to independent antibody HPA038848 (B).
Orthogonal Validation Using RNA-Seq
Orthogonal Validation Using RNA-seq - Demonstrating antibody specificity through an antibody-dependent method correlated with an antibody-independent method (RNA-seq).
Expected Results: Antibody staining intensities should correlate to RNA-seq data from the same samples.
In the antibody validation process, the practice of orthogonal validation refers to the comparison of immunodetection results with a method that is antibody-independent for measuring the gene or protein of interest.
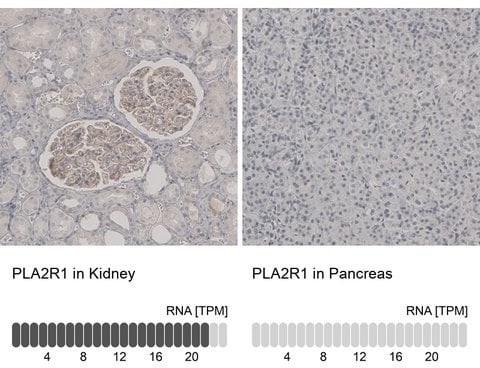
The orthogonal technique, RNA-seq, is a method used to quantify gene expression at the mRNA level for a given sample for a specific target. This expression at the mRNA level should correspond to protein expression. For this pillar of enhanced antibody validation, data is presented demonstrating the mRNA expression level obtained by RNA-seq for a given target in bar graph form for a high- and a low-expressing tissue (5-fold difference) alongside WB or IHC data for those same samples. What is demonstrated is a direct comparison of mRNA expression level and sample staining: High expression=strong staining; Low expression=weak or no staining.
Orthogonal validation using RNAseq.
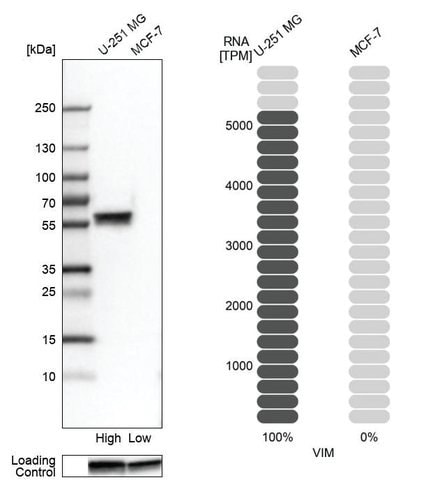
Orthogonal validation in Western blot (left) and IHC (right). Samples with known high and low RNA expression were chosen. Left: Western blot analysis (left) using an anti-VIM antibody in human cell lines U-251MG (high expression of VIM) and MCF-7 (low expression of VIM) correlates with RNA-seq data (right).
Right: Immunohistochemistry analysis using an anti-PLA2R1 antibody in human kidney (left, high expression of PLA2R1) and pancreas tissues (right, low expression of PLA2R1) correlates with RNA-seq data.
Functional Assay Validation
Functional Assay Validation - Demonstrating antibody specificity through immunodetection of experimentally induced changes in target antigen expression and or activity levels.
Expected Results: Immunodetection of altered antigen activity or expression levels via WB, ICC, IF, and FS.
Many antigen expression and activity levels can be readably modulated in vitro by experimental manipulation. Leveraging common protein expression or bioactivity-altering techniques allows for the determination of antibody specificity by detecting changes in the activation or expression levels of the target protein. Functional assay validation is a valuable EV pillar as it provides strong supporting evidence of antibody specificity.
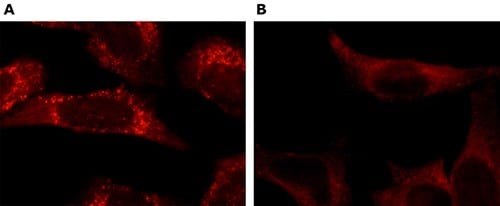
Immunofluorescence
For the functional assay validation pillar, HeLa cells were incubated in EBSS medium for 2 hr and autophagosome staining is demonstrated in the experimentally induced HeLa cells (A) while no autophagosome staining is demonstrated in the untreated cells (B). Cells were fixed and permeabilized with cold methanol followed by cold acetone and stained with 5 μg/mL Rabbit Anti-LC3B (Cat. No. L7543), a marker for autophagosome membranes. The antibody was developed using Goat Anti-Rabbit IgG, Cy3 conjugate.
Our steadfast commitment to antibodies validated for life science research
We are a leading provider of primary antibody products that are validated every day by your peers through the immense number of peer reviewed citations of the use of our antibodies. Industry leading quality control processes and application testing ensure that you can rely on our antibodies to provide the expected performance the first time and every time. In addition to our long-standing commitment to validation, we continue to expand and improve our processes by incorporating more rigorous validation strategies into the development of many of our antibody reagents. We stand by the quality of our product with our Antibody Guarantee—if our antibody does not perform as specified in your application, we will issue a full credit or replacement antibody product.
Expression/Overexpression Validation
Expression/overexpression Validation - Demonstrating antibody specificity through overexpression of the target protein.
Expected Results: Immunodetection of the overexpressed protein via WB and IF.
The expression/overexpression validation ensures specificity of the antibody using transfection to overexpress the target antigen, preferably in a cell line that does not already express the protein. The overexpressed protein condition is compared to a wild type control sample that has been demonstrated to not express the target protein. The specificity of the antibody is subsequently assessed through WB or IF staining of the cell sample that has been altered to overexpress the target protein.
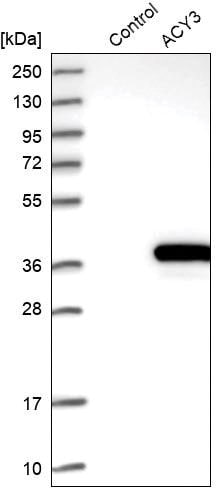
Enhanced validation by overexpression of the target protein
Antibody recognition
(Anti-ACY3 antibody
produced in rabbit)
is confirmed by western blot in the cell sample that has been altered to overexpress the target protein while no staining is detected in the wild type sample (control).
Immunocapture Followed by Mass Spectrometry (MS) Validation
Immunocapture Followed by Mass Spectrometry (MS) Validation - Demonstrating antibody specificity using the antibody immunocapture technique followed by MS analysis.
Expected Results: MS analyses confirm that the antibody is directly interacting with the target protein. Immunocapture utilizes immunoprecipitation (IP) to isolate and capture the antibody that is bound to the target protein from the cell lystate. Following IP, MS analysis confirms that the antibody is directly interacting with the target protein by identification of the target protein peptide sequences.
References
For Research Use Only. Not for use in diagnostic procedures.
Unless otherwise stated in the Product(s) specifications, any Antibody product is sold for internal research use only and may not be used for any other purpose, which includes but is not limited to, any commercial, diagnostic, or therapeutic use. Our validation processes pertain only to research uses and do not confirm or assure that our antibodies can be used for any unauthorized uses as set forth herein.
To continue reading please sign in or create an account.
Don't Have An Account?