Epigenetic Mechanisms in Cancer
Cancer accounted for approximately 13% of all deaths in 2004, according to the cancer fact sheet published by the World Health Organization (WHO) in February 2009. The term cancer (malignancy) describes a diverse class of diseases characterized by uncontrolled cell growth, which frequently culminates in the invasion into and the destruction of adjacent tissue - and subsequent metastasis to other locations in the body via lymph or blood. It is these malignant properties that differentiate cancer from normal cells and benign tumors that tend to be self-limited, noninvasive and do not metastasize.
Nearly all cancers arise gradually, resulting from the accumulation of abnormalities (mutations) in the cell's genetic material due to the effects of carcinogens, ionizing radiation, viral infection, errors in deoxyribonucleic acid (DNA) replication, or inherited defects. This accumulation of mutations in cells contributes to the invasive and metastatic properties of cancer cells as well as their ability to take up residence in an array of tissue environments. Genetic abnormalities resulting in cancer frequently occur in oncogenes (genes that cause the transformation of normal cells into cancerous tumor cells) or in tumor suppressor genes.
Proto-oncogenes (e.g., ras, wnt, myc and erk) code for proteins that help to regulate cell growth and differentiation and are often involved in the signal transduction and the execution of mitogenic signals. Mutations in these genes result in their activation and lead to hyperactive cell growth and division, or a newfound protection against apoptosis. An additional cause of cancer results from mutations occurring in tumor suppressors such as p53 and the retinoblastoma protein. The down-regulation of these proteins has a dampening or repressive effect on the regulation of the cell cycle or promotes apoptosis, and sometimes does both.1 Usually in combination with other genetic changes, these mutations result in a loss or reduction in gene function and enable the cell to progress towards cancer.
Identifying Candidate Genes for Activity in Cancer Progression
While the study of the whole organism often provides the most data, it is also complex due to the interactions between tissue types and the organism's endocrine and paracrine pathways. Many stages of the progression of cancer can also be modeled at the cell-based level, allowing the elucidation of the genetic mechanisms underneath many of the overlying layers of cell-type interaction. There is a large number of molecular interactions that lead to the causal phenotype within a given system, pathway, or disease. To better understand this complexity, screening to identify the major candidates causally involved in these pathways often lies at the beginning of most discovery research. Researchers are now moving toward large screening projects, which are often genome-wide studies, to identify the candidate genes involved in the biological question of interest. Discovering these biological interactions leads to a host of characterization and validation studies requiring further evaluation.
The flexibility of its applications has made ribonucleic acid interference (RNAi) a valuable tool for researchers who are interested in gene function characterization, signaling pathway analysis and drug target validation. RNAi utilizes a sequence-specific inhibition of gene expression to allow a targeted approach to identify the role of gene activity, based on the resulting loss-offunction phenotypes. The ease of use allows the generation of whole genome screening tools. Both short interfering RNA (siRNA) and short hairpin RNA (shRNA) platforms have been utilized in screening. The primary advantages of using synthetic siRNAs for screening are found in their ease of use and the greater availability of established delivery methods. Available siRNA libraries targeting nearly every annotated gene have been extensively validated and provide robust gene silencing.2
More recently, an alternative RNAi pooling method employing enzymatically synthesized RNA (esiRNAs) (Figure 1), has been used in whole genome screening in mammalian cells.3 The initial discovery of esiRNAs took place in the laboratory of Michael J. Bishop and was subsequently developed by Frank Bucholz for screening applications at the Max Planck Institute in Germany. This technology involves the generation of endoribonucleaseprepared siRNAs (esiRNAs) by the in vitro transcription of a 300-600 bp gene specific double-stranded RNA (dsRNA), followed by enzymatic digestion using ribonucleases (RNases) such as RNase III. This digest produces complex pools of siRNA-like molecules, and these multiple silencing triggers lead to specific and effective gene silencing.
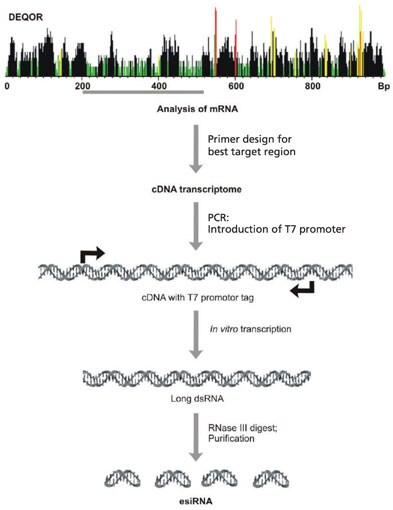
Figure 1.A cDNA transcriptome is generated from a region of mRNA. The region to be amplified is selected based on the higest possible number of highly effective siRNA based on the Deqor siRNA design program. PCR is used to introduce T7 promoters for the in vitro transcription of long dsRNA. Finally, the long dsRNA is enzymatically digested to short dsRNAs producing complex pools of siRNA-like molecules. The digestion is then purified to remove any remaining DNA template, unincorporated nucleotides, and dsRNAs longer than approximately 48 bp.
For stable long-term silencing, shRNAs are the RNAi platform of choice for complex pathway analysis and screening. Delivering the shRNAs via a lentiviral particle provides numerous features that are useful for screening studies as well, including broad tropism, stable integration, and negligible interferon response upon transduction. With lentiviral delivery, the shRNA sequence stably integrates into the host chromosome for the long-term reduction of messenger RNA (mRNA), thereby extending the assay time and allowing for the measurement of protein reduction.
The RNAi Consortium (TRC), a public/private consortium led by the Broad Institute of MIT and Harvard, has developed lentiviral-based shRNA libraries that provide coverage of the murine and human genomes, and these libraries have been used in a number of screens.4 A variety of RNAi screening tools based on the TRC genome-wide libraries are now commercially available, including both pooled or pre-arrayed, to allow for more flexible methods to screen whole genomes.
These processes have become increasingly significant in drug R&D, and RNAi can be used to facilitate the identification and the validation of drug targets. In cancer research, RNAi is useful for characterizing molecular targets, screening genes, and looking at gene interactions with specific drugs - indispensable to the field of pharmacogenomics.
Epigenetics and Cancer
Although genetic lesions have been the focus of cancer research for many years, it has been increasingly recognized that aberrant epigenetic modifications also play a major role in tumorigenesis. Epigenetic changes, referring to heritable changes in gene expression occurring without alteration in a DNA sequence, contribute to the pathogenesis of cancer by altering gene expression.5 Furthermore, both genetics and epigenetics cooperate at all stages of cancer development. While the accumulation of genetic lesions and aberrant epigenetic regulation are causative agents in the formation of cancer, RNA (in particular microRNA or miRNA) is a final player that necessitates mention. MicroRNA comprises a network that controls gene expression and protein production throughout the body and is intimately involved in the formation of a repressive chromatin state.6 Scientists believe miRNAs play a major role in controlling overall gene expression and the cancerous process.
Epigenetics refers to all heritable modifications to genes other than changes in the DNA sequence itself. The Greek prefix "epi-" implies features that are on top of, or in addition to genetics. Epigenetic modifications influence the appearance and structure of DNA and regulate gene expression. One example of an epigenetic mechanism, DNA methylation, refers to the addition of a methyl group to cytosine in a cytosine-phosphate-guanine (CpG) dinucleotide.
Once a cell has an established DNA methylation pattern, these sites are inherited by daughter cells and can have important implications in normal cellular function and development. Additionally, aberrant epigenetic modifications play a major role in the development of cancer and other conditions in which cell and tissue growth are abnormal. Incorrect epigenetic changes to tumor suppressor genes and oncogenes are some of the first steps in cancer initiation. In cancer, some tumor suppressor genes are mistakenly turned off, preventing the growthlimiting protein from being made. Likewise, many oncogenes, or growth-promoting genes, are mistakenly turned on, resulting in abnormal cell proliferation.
In cancer cells, genes can be modified by mutations, which alter the function of the proteins that the genes encode. Through epigenetics, modifications to chromosomes take place and alter gene-expression patterns. Epigenetics changes occur through DNA methylation, and the methylation, acetylation, or phosphorylation (the addition of a phosphate to a protein or an organic molecule) of histones and other proteins around which DNA is wound, to form chromatin. The theory of epigenetics in cancer pathogenesis is that non-mutational changes to DNA lead to alterations in gene expression. In a normal cell for example, an oncogene may be silent as a result of DNA methylation, and the loss of that methylation may induce the aberrant expression of a gene, contributing to cancer progression. Likewise, tumor suppressor genes may be silenced by DNA hypermethylation during cancer development. Current epigenetic therapy takes advantage of the reversibility of these "epimutations".
Both genetics and epigenetics cooperate at all stages of cancer development. To date, most of the mutations that have been identified as contributors to cancer formation are associated with tumor initiation. In contrast, few specific genetic mutations have been linked to tumor progression, suggesting that epigenetic changes may be involved. In this scenario, a genetic mutation initiates the cancer but epigenetic changes including DNA methylation and histone modification promote its progression. However, this does not mean that epigenetic changes do not participate in cancer initiation as well. For instance, a decrease in methylation can result in the loss of imprinting to genes that participate in the regulation of growth, which can lead to cancer. Furthermore, changes within the epigenome may "prime" cells in such a way as to promote cellular transformation upon a subsequent DNA mutagenic event.
DNA Methylation
DNA methylation is an essential part of normal development and is associated with imprinting, X-chromosome inactivation, suppression of repetitive elements, and carcinogenesis. DNA methylation involves the addition of a methyl group to DNA, for example, to the number-5 carbon of the cytosine pyrimidine ring. Like other epigenetic mechanisms, it is heritable and does not alter the underlying DNA sequence. In humans, DNA methylation occurs at CpG dinucleotides and 60-90% of CpG sequences are methylated in the genome of adult somatic tissue. The maintenance methylation activity catalyzed by DNA methyltransferase (DNMT) preserves DNA methylation on daughter strands after DNA replication.
Methylation is repressive to transcription and serves to enhance genome stability. DNA methylation impacts gene transcription in two ways. First, the methylation of DNA physically impedes the binding of transcriptional proteins to the gene and secondly, methylated DNA is often bound by methyl-CpG-binding domain (MBD) proteins. MBD proteins recruit additional proteins, including chromatin-remodeling proteins such as histone methyltransferases (HMTs) and histone deacetylases (HDACs) to the locus. The net result is compact, inactive chromatin.
DNA methylation was the first epigenetic alteration to be observed in cancer cells.7 The 5' regulatory regions (promoters) of many genes contain "CpG islands" that escape DNA methylation (Figure 2). In some cancers, these CpG islands acquire abnormal hypermethylation, resulting in heritable transcriptional silencing by this process. The reverse is also true as some CpG islands that are normally methylated acquire abnormal hypomethylation resulting in gene activation.
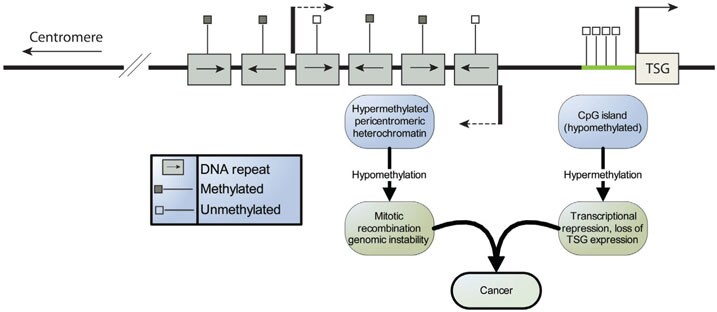
Figure 2.DNA Methylation and Cancer. This region of genomic DNA contains hypermethylated heterochromatin and an actively transcribed Tumor Suppressor Gene (TSG). A hallmark of tumor cells is the conversion of repeat rich regions from hypermethylated to hypomethylated, culminating in genomic instability resulting from increased mitotic recombination. Additionally, the CpG island located upstream of the TSG is hypomethylated, allowing for transcription. However, during oncogenesis, the CpG islands become hypermethylated, resulting in a loss of expression and a progression toward cancer.
A general tendency affiliated with aging is for the genome to become hypomethylated while some CpG islands become hypermethylated. The net result is an increase in the incidence of cancer with age. Based on what is known about methylation, it is easy to appreciate how this process may facilitate tumorigenesis. For example, the hypermethylation of CpG islands in tumor suppressor genes switches off these genes, whereas global hypomethylation promotes genomic instability and the subsequent activation of oncogenes and transposable elements (transposons are mutagens).
DNA methylation is also the principal epigenetic factor governing allelic imprinting. Genomic imprinting is a genetic phenomenon by which certain genes are expressed in a parent-of-origin-specific manner independent of classical Mendelian inheritance. For the majority of autosomal genes (i.e., genes found on any of the chromosomes other than the gender-determining chromosomes), expression occurs from both alleles simultaneously. However, a small proportion (<1%) of genes are imprinted and expression occurs from only one allele.
Imprinted genes are either expressed only from the allele inherited from the mother (e.g., H19 or CDKN1C) or in other instances from the allele inherited from the father (e.g., IGF2). The regulation of imprinted genes is largely dependent on methylation marks laid down during the embryological development of germ cells. The stability of these methylated regions in somatic cells is maintained through each cellular replication by DNMT1. The aberrant regulation of imprinted gene expression (Loss of Imprinting or LOI) is seen frequently in a variety of human tumors, and may be considered the most abundant and precocious alteration in cancer.8 There are two primary outcomes resulting from LOI:
- The activation of a normally silent copy of a growth-promoting gene, such as the insulin-like growth factor-2 (IGF2), or
- The silencing of the normally active copy of a growth-inhibitory gene, such as p57 KIP2.
Medical scientists are studying DNA methylation and human disease to determine the connections between methylation abnormalities, gene expression, and silencing. They are also examining various diseases such as cancer, lupus, muscular dystrophy, and a range of birth defects that appear to be caused by defective imprinting mechanisms.
Hypomethylating Drugs
According to Gronbaek, the current approach to cancer treatment - a direct result of hypermethylation - is to employ hypomethylating drugs.5 For example, the drug Dacogen® (5-aza-CdR) has been approved by the U.S. Food and Drug Administration (FDA) for use in the treatment of certain hematopoietic cancers. When this deoxycytidine analogue is incorporated into DNA, it covalently binds to DNMTs, sequestering the enzymes. Consequently, DNMT is no longer available for maintaining the methylation pattern in newly synthesized DNA strands during replication, resulting in the successive loss of the methylation signature. More importantly, the demethylation activity occurs in actively dividing cells. Because the growth rate of cancer cells is typically higher than that of normal cells, this activity does not affect the epigenetic silencing patterns of normal cells, which is important as the demethylation of oncogenes in normal cells would lead to cancer.
Hypomethylating Drugs
Eukaryotic genes are located on multiple linear chromosomes that are packed into a complex with histone proteins to form chromatin. The structure of chromatin is dynamic, existing in either a heterochromatin (condensed) or euchromatin (extended) state. The basic unit of chromatin is the nucleosome, which is folded through a series of successively higher order structures to eventually form a chromosome. Nucleosomes are composed of 146 bp of DNA around a histone octamer, consisting of two copies each of the core histones H2A, H2B, H3, and H4 with a linker H1. In the absence of linker histones, the nucleosome adopts a less condensed, or "relaxed" form of chromatin, commonly referred to as "Beads-on-a-String".
The overexpression or mutation of HAT genes is a component of a variety of cancers, especially those of hematological and epithelial origin. For example, the p300 HAT gene is mutated in a number of gastrointestinal tumors. Changes affecting the normal function of HDAC genes as a causative agent in cancer formation are far less common. However, the aberrant targeting of HDACs is associated with the transcriptional silencing of tumor suppressor genes, including the cyclin dependent kinase inhibitor p21 that blocks cell cycle progression.
In this example, histones are improperly deacetylated and bind more tightly to the regulatory elements of the p21 gene, preventing the latter's transcription and resulting in the inhibition of the p21 expression. The outcome is uncontrolled cell division. HDAC inhibitors can reactivate the p21 expression and thereby prevent tumor cell proliferation. HDAC inhibitors are performing well in the clinic as anti-cancer drugs and have significant anti-tumor activity.9
Histone methylation is more complex than acetylation as both lysine and arginine residues can be methylated on side-chain nitrogen atoms by histone methyltransferases, and one or more methyl groups can methylate each amino acid residue. This complexity provides regulatory potential as each event may affect the chromatin structure and the ability to interact with regulatory proteins. Histone methylation is generally associated with transcriptional repression. However, the methylation of some lysine and arginine residues of histones results in transcriptional activation. Examples include methylation of lysine (K) 4 of histone 3 (H3K4), and arginine (R) residues on H3 and H4 (Figure 3).
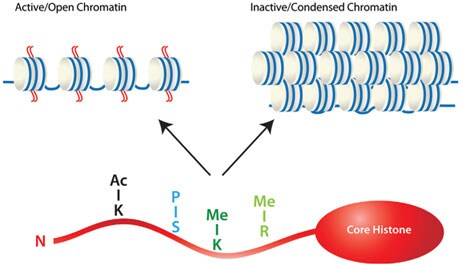
Figure 3.Eukaryotic genes are located on multiple linear chromosomes packed into a complex with histone proteins to form chromatin. Histones are the primary regulator of the chromatin structure, which is dynamic, existing in either a heterochromatin (condensed) or euchromatin (open) state. Whether chromatin is active or inactive is largely dependent on the post-transcriptional modifications of the histone tails, including acetylation, methylation, and phosphorylation.
The Supressor of variegation-Enhanser of zeste-Trithorax (SET) domain protein methyltransferase superfamily methylates histones on lysine, and numerous SET domain-containing proteins are implicated in cancer. For example, SUV39 is a SET domain-containing methyltransferase that catalyzes the methylation of K9 in H3. Mice that carry deletions of SUV39 and its family members suffer genome instability that is associated with a substantial loss of H3K9 methylation. Transgenic mice devoid of this enzyme are susceptible to cancer, especially B cell lymphomas.
Several histones are subject to phosphorylation, and this modification is associated with large-scale chromatin reorganization during processes such as mitosis, apoptosis, and DNA repair. Phosphorylation of the linker histone H1 by CDK2 is associated with cell-cycle progression, and H1 phosphorylation has long been a marker for mitotic cells. However, while elevated H1 phosphorylation is observed in many cancer cells, this may be an artifact of the enhanced proliferation of these cells. Nevertheless, the improper regulation of the kinases mediating histone phosphorylation can be oncogenic. For example, the Aurora kinases normally phosphorylate H3 and control a number of mitotic (cell division) events. All three family members (Aurora A, B, and C) are overexpressed in many aggressive human cancers, resulting in defective chromosome segregation and aneuploidy.
Studying the Epigenome
Epigenetic gene silencing is a major driving force in cancer, and the study of epigenetic mechanisms in the disease, such as DNA methylation, histone modifications, and micro-RNA expression, have revealed factors that contribute to the neoplastic phenotype. The methylation of cytosine in promoter regions and the covalent modification of chromatin proteins such as histones cause the suppression of transcription. Many of these silenced genes are tumor suppressors, and promoter/CpG island hypermethylation is frequently associated with poor disease prognosis. Understanding the trigger for these epigenetic changes will be essential to reduce or prevent them from occurring. DNA methylation may be the best characterized epigenetic process, largely due to the fact that it is the easiest to study with existing technology. Methods for DNA methylation analysis can be roughly divided into two types: global and gene-specific methylation analysis.
The overall methylation status of a genome can be a useful measure of global regulatory changes. Methods for measuring global methylation include high-performance liquid chromatography (HPLC), mass spectrometry, and methyl accepting capacity assays. These methods measure the overall level of methyl cytosines in the genome. Enzyme-linked immunosorbent assays (ELISA) are a fourth method for determining global methylation. Some commercially available kits utilize a sandwich ELISA-based method to quantify methylated DNA colorimetrically. The amount of methylated DNA present in the sample is proportional to the absorbance measured. The advantage of this method over HPLC and mass spectrometry is that the format is easier to implement in the lab.
A number of techniques have been developed for gene-specific methylation analysis. Most early studies used methylation sensitive restriction enzymes to digest DNA, followed by Southern detection or polymerase chain reaction (PCR) amplification. Bisulfite reactionbased methods have become popular, such as methylation specific PCR (MSP) and bisulfite genomic sequencing PCR. Methylated cytosine (meC) is stable with bisulfite while cytosine is converted to uracil. Therefore, bisulfite treatment is employed to introduce specific changes in the DNA sequence that depend on the methylation status of individual cytosine residues. There are numerous commercially available kits containing the necessary components to treat DNA with bisulfite to convert cytosine residues to uracil, while leaving 5-methylcytosine residues unaffected. Converted DNA is suitable for a variety of downstream applications including MSP, methylation sequencing, and pyrosequencing. Additionally, genome-wide screen methods have been developed, such as restriction landmark genomic scanning for methylation (RLGS-M) and CpG island microarray to identify unknown methylation hot-spots or methylated CpG islands in the genome.
Another significant epigenetic process is chromatin modification. Histone modifications change the chromatin structure, and are an early indicator of epigenetic regulation. One way to study this phenomenon is via chromatin immunoprecipitation (ChIP). This technique starts with cells, and uses a cross-linking agent to chemically link the DNA and the latter's interacting proteins. The resulting DNA is isolated, sheared, and precipitated from the bulk, using a protein specific antibody (e.g., acetylated histone). The cross-links are reversed, and the precipitated DNA, now enriched for sequences that interact with the protein of interest, is examined to determine which sequences are present. Detection can be via PCR when looking for a few genes or can be performed using microarrays (ChIP-chip) or parallel (deep) sequencing (ChIP-sequencing). There are commercially available kits on the market for ChIP analysis including kits using a 96 well format for high throughput analyses. ChIP-chip requires the amplification of the enriched DNA sample, as immunoprecipitation does not supply the amount of DNA that is required for microarray analysis. Whole genome amplification has been successfully applied to ChIP DNA amplification, and is a method for generating more DNA from a fragmented DNA sample.10
Discovery and Validation of Epigenetic Drugs
Unlike genetic alterations, epigenetic changes are potentially reversible and the large-scale development of small molecule inhibitors of DNA and histone-modifying enzymes is in full swing. The utilization of epigenetic targets is emerging as an effective and valuable approach to chemotherapy as well as the chemoprevention of cancer. The success of HDAC inhibitors and DNA demethylating agents like 5-aza-CdR as anti-cancer drugs, demonstrates proof-of-principle of this approach and provides hope for the development of a more comprehensive portfolio of "epigenetic drugs" in the future.
Until a decade ago, research programs were focused on identifying and quantifying the environmental and inherited factors that are associated with cancers. By utilizing improved screening tools such as RNAi and small molecules, a basic understanding of the mechanisms has been attained as well as a glimpse into the increasingly intricate interactions between genes. However, it is increasingly being recognized that aberrant epigenetic modifications also play a major role in tumorigenesis. More than 600 genes have been identified that are regulated by epigenetic mechanisms, including tumor suppressor genes, oncogenes, and cancerassociated viral genes.
Epigenetic mechanisms driving cancer formation include changes in primary DNA methylation, modifications to the histone code related to DNA methylation, and abnormalities in specific histone modifying enzymes. The field is rapidly moving towards clinical applications for the treatment of patients with cancer or evaluating a patient's level of risk for developing cancer. Assays for aberrant DNA methylation, bisulfite modification, high-throughput bisulfite genomic sequencing, quantifying global DNA methylation, combinations of histone deacetylase inhibitors and DNA methyltransferase inhibition assays, chromoimmunoprecipitation and RNAi interference, address the role of epigenetics in cancer.
With next generation genomic platforms and methods of screening whole genomes and the subsequent analysis of drug target candidates, scientists are able to cost-effectively assay individual cancer genomes. These genomes can be characterized in terms of global genetic, epigenetic, and transcriptional changes. The development of drug discovery tools like RNAi and continued improvements in high-throughput robotics, data processing, control software, and liquid handling devices will accelerate this process. In-depth characterization of these events and the interdependent relationships between them will lead to a better understanding of the mechanisms of tumorigenesis, metastasis, and therapeutic response.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?