Yeast Transformation Protocols
Introduction
Yeasts are considered model systems for eukaryotic studies as they exhibit fast growth and have dispersed cells. Moreover, replica plating and mutant isolation of yeast cells can be done with relative ease and they have a well-defined genetic system. Most significantly, yeasts have a highly versatile DNA transformation system that can be utilized effectively for protein production.
Yeast Transformation
Yeast Transformation Protocol
Protocol Overview
The LiAc transformation method involves three main steps: preparing competent yeast cells, transformation with plasmid DNA, and subsequent plating to select the transformants. Yeast transformants are usually selected using auxotrophic markers; hence, the appropriate synthetic dropout medium is used for screening transformants.
Reagents Required:
Stock Solutions
50 % Polyethylene glycol solution (mol wt ~36,500)
1 M Tris-HCl and 0.2 M EDTA, pH 8.0 (TE Buffer) (Catalog Number T9285)
1 M Lithium acetate, pH 7.5 (Catalog Number L4158)
1x TE-LiAc solution
10 mM Tris-HCl, pH 8.0, with 1 mM EDTA and 0.1 M lithium acetate
PEG-TE-LiAc solution
40 % PEG in 10 mM Tris-HCl, pH 8.0, with 1 mM EDTA and 0.1 M Lithium actetate
Yeast Transformation
- Prepare YPD and synthetic complete (SC) drop-out medium plates and autoclave them separately (Tables 1 and 2 ).
- Inoculate yeast cells from plates into 20 mL of YPD medium in a 100 mL sterile flask.
- Grow overnight with shaking.
- Dilute cells from above culture into 100 mL of YPD medium until the OD600 is 0.3
- Pellet cells gently.
- Resuspend in 7-8 mL of 1x TE-LiAc solution and rotate at 23 °C for 1-1.5 hours.
- Add 10 µL of 10 mg/mL salmon testes DNA (Catalog Number D9156) in sterile microfuge tubes designated for transformation and one for a negative control.
- Add 0.1 µg of yeast plasmid DNA (to be studied) to each tube and 100 µL of competent cells into each tube and then vortex.
- Add 600 µL of freshly prepared PEG-TE-LiAc solution, vortex, and incubate at 30 °C for 30 minutes with shaking.
- Optional - DMSO (Catalog Number D8418) can be added to 10% (v/v); followed by heat shock for 15 minutes at 42 °C.
- Spin for 3 seconds, resuspend cells in sterile water and plate using appropriate SC drop-out medium.
Note: Refer to growth protocols for plating yeasts and the following section for isolating transformants.
Isolation of Yeast Transformants
Yeast transformants are usually selected using the URA3 complementation method, although techniques utilizing amino acids for complementation and blue-white screening methods can also be used.
In the URA3-based selection technique; the plasmid DNA has a normal copy of the yeast URA3 gene, as well as the URA3 promoter; whereas, the yeast mutant strain lacks the URA3 allele. Thus, yeast cells transformed with the plasmid have a functional copy of the URA3 gene, which allows them to grow on Yeast Synthetic Drop-out Medium Supplements without Uracil (Catalog Number Y1501).
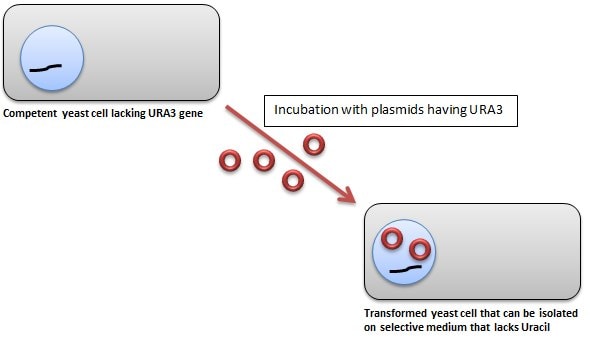
Figure 1.Yeast Transformation
Materials
References
To continue reading please sign in or create an account.
Don't Have An Account?