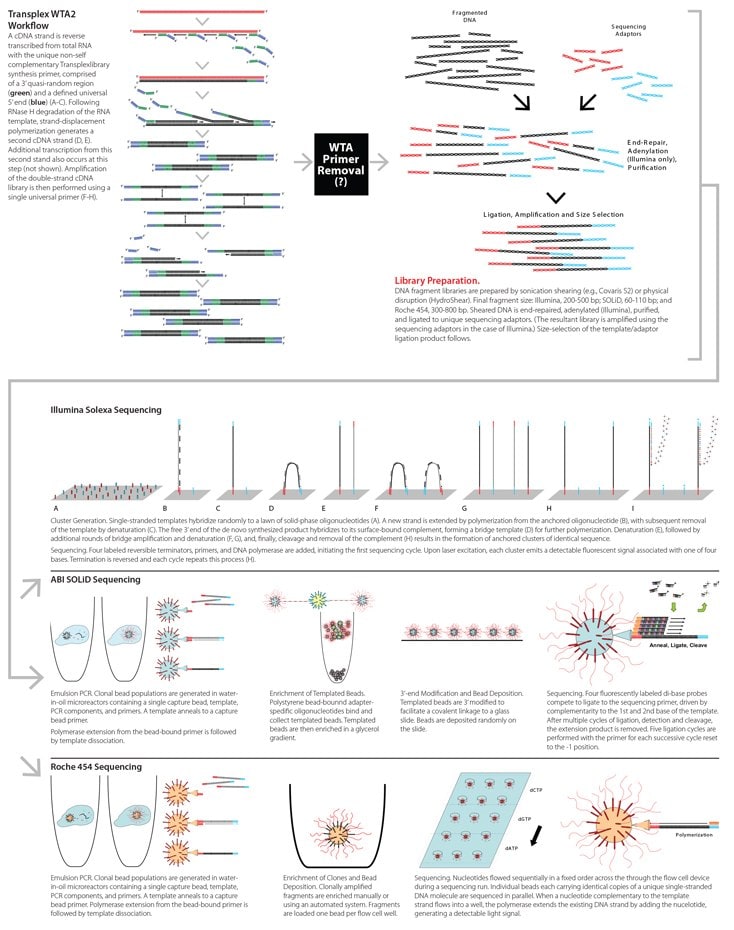
WTA2 Amplicon Modification for NGS
Since the publication of this article, we have launched a WTA2 NGS compatible kit, SEQR, that includes primer removal.
Abstract
Whole Transcriptome Amplification (WTA2) exponentially amplifies RNA producing a double-stranded cDNA library while precisely maintaining differential levels of individual transcripts in test and reference samples. Though originally designed to amplify nanogram quantities of RNA, WTA2 has been shown to be exceedingly effective for amplification from damaged RNA template (FFPE and laser captured tissue samples) and single-cell input quantities (picograms). The efficacy of WTA2 amplification for downstream applications, primarily qPCR and expression microarray analysis, is well-documented. It follows that the utilization of next generation sequencing for gene expression research and diagnostics would be well served by amplification of RNA isolated from samples of severely restricted quantity or quality.
Strategies for the integration of WTA2 with next-generation sequencing are examined, with particular emphasis on elimination of the characteristic fixed primer sequence associated with each amplicon in the amplification library. Removal of these sites will allow direct entry of the resulting product into the sequencing workflow. Methods under consideration will enable the WTA2 amplicon to feed into the current sample prep protocols for the Illumina GA and GAII, SoLiD 5500/5500xl, and Roche-454 GS FLX/Junior platforms.
Introduction
Platform Strengths
Complete WTA2 kit has been demonstrated to effectively amplify damaged and/or low quantities of RNA template1-13. Exponential PCR-based amplification provides both sensitivity and reproducibility, while maintaining relative levels of transcript abundance1,2,4,8,10-12.
- The WTA2 library synthesis primer design was improved to provide increased coverage of the transcriptome (over that of WTA1), allowing for enhanced ability to amplify fragmented RNA14.
- The 3’ component of library synthesis primer, comprised of quasi-random sequence (green, WTA2 Workflow), primes throughout the entire length of its respective template during first- and second strand cDNA synthesis, eliminating the 3’ bias15,16 characteristic of the “Eberwine” amplification methods17. Hence, WTA2 amplicon is perfectly suited for exon and alternative splicing studies.
- The single universal primer (blue, WTA2 Workflow) is non-self complementary, eliminating potential intra- and intermolecular hybridization of the primer itself18. This, combined with amplification conditions and reaction parameters that help to prevent intra-molecular hybridization of the complementary terminal ends of each amplicon during amplification19, provides for substantially better amplification efficiency over platforms the rely on random “dN” sequence for library synthesis.
Misconceptions
The most common misconception, detracting from the general acceptance of PCR-based amplification strategies, is the comparison of the disjointed increase in relative levels of two different transcripts during amplification in the same reaction—due primarily to differences in amplicon length and complexity. This is not a legitimate argument. WTA2 amplification technology addresses this misconception through the following points:
- The imperative for RNA amplification is the maintenance of relative levels of the same amplicon in, minimally, two samples: the test and reference sample. All amplification methods possess systematic bias, and therefore differential expression, not individual reads, is the true standard. In this way, WTA is highly reproducible1,2,4.
- Considering the expression differential between the test and reference sample, PCR amplification is more sensitive, able, to detect stochastic basal expression (eliminated from consideration by statistical analyses)4.
Applications
- Recent publications have demonstrated the proficiency of the WTA2 kit for microarray application, including rigorous evaluations of the amplification method, in the case of Gonzalez-Roca et al4, and Gilbert et al8 and Robert16.
- In addition, Nimblegen and Agilent microarray platform providers have evaluated and recommend WTA210-12.
- Procedures for integration of WTA2 and the popular microarray platform workflows, and for downstream qPCR application are provided on the Sigma-Aldrich website20.
- Though not reported in the literature, WTA2 can serve as a vital “archiving” tool, storing RNA sequence information as stable cDNA for future alternative analytical methods.
Modification of Amplified RNA for NGS Sequencing
The non-self complementary primer sequence is the linchpin of amplification, critical for efficient, positionally unbiased amplification of each transcript and comprehensive coverage of the transcriptome. The 5’ universal sequence spans 22 nucleotides, with the quasi-random 3’ region comprising 10 nucleotides (WTA2 Workflow). Because of the significant cost of labeled nucleotides and characteristic short read lengths for the Illumina and ABI platforms (Table 1), it is cost-effective to remove these primer sequences prior to NGS sequencing.
Several enzymatic processes for primer sequence removal are under consideration.
- Incorporation of a type II restriction site in the universal primer sequence. This would allow for cleavage of both universal and quasi-random primer sequence from the amplified cDNA prior to sequencing adaptor ligation (Library Preparation).
- Amplification incorporating modified dNTPs. Random incorporation of modified dNTPs during amplification is followed by nuclease digestion and 3’ end polishing (and dA tailing as required). This approach will statistically remove a majority of WTA library and amplification primer-derived sequences23 prior to sequencing adaptor ligation (Library Preparation).
Click on image below for larger view.
An alternative to enzymatic manipulation of the WTA2-amplified library is the following.
- Sequencing oligonucleotide adaptors are ligated to the WTA2 amplicon (without removal of any primer sequence), as required by the respective sequencing platform workflow (Library Preparation).
- The platform sequencing primer would be replaced by an alternative primer complementary to the WTA2 universal amplification primer (Sequencing: Illumina, Roche 454).
- This approach will eliminate the amplification primer sequence from further analysis, but leave the quasi-random library synthesis sequence.
Discussion
The expectations of researchers are driving further development of methodologies and technology for single-cell analyses. Examples of RNA-Seq24-26 and RIP-Seq27 analyses (RNA Immunoprecipitation) are becoming more prevalent, in the study of disease and development, particularly in the assessment of oocyte and embryo development28.
Challenges for NGS Sequencing
- Cell expression profiling requires robust methods of RNA amplification for generation of sufficient starting material for deep sequencing29 (Table 2).
WTA2 produces 3 to 5 µg of amplified product from 20 picograms of input total RNA (a singlecell equivalent quantity).
- Normalization between samples must account for expression level, gene length of the genes, and the complexity of the RNA population33.
Conclusions
- Complete WTA2 kit has been demonstrated to be the method of choice for amplification of damaged and/or low quantities of RNA for microarray application.
- Efforts are underway to modify the WTA2 amplicon to allow for easy and efficient entry into workflows of the current popular deep sequencing platforms.
Materials
To continue reading please sign in or create an account.
Don't Have An Account?