Fragment-Based DNA-Encoded Library for Drug Discovery
Accelerating Early Drug Screening Efforts
DNA-Encoded Library (DEL) technology is an increasingly mature and prominent approach to early-stage drug discovery. As opposed to traditional high-throughput screening approaches where compounds are tested individually over months in a target-specific assay, DEL screening allows huge numbers of compounds to be simultaneously interrogated in a largely target-independent binding assay.
The principle is straightforward: organic molecules are conjugated to unique DNA strands which function as identifying barcodes. The target protein is attached to a solid support and treated with the library. After washing off non-binding compounds, the identities of hit compounds can be revealed via DNA sequencing. These medicinal chemistry starting points can then be investigated in follow-up experiments1.
Accessible, Reliable DNA-Encoded Library Technology
DyNAbind and Merck KGaA, Darmstadt, Germany have joined forces to make a fragment-based DEL kit available to researchers working on innovative protein targets. We’ve selected over 370,000 fragment pairs to cover a broad range of druglike chemical space and cast the widest possible net for your project.
If you felt previously priced out of large library drug discovery, this is the kit for you. Any group with common biochemistry and molecular biology equipment and skills can set up, run and prepare the kit for sequencing.
The library is built on DyNAbind’s proprietary Dynamic Library architecture. This novel DNA assembly permits dynamic reshuffling of the library members, until they’re stabilized by binding to the target protein. The result is over 30-fold increased signal-to-noise ratios, for more clear and trustworthy results2,3.
Designed for MedChem Relevance
An effective drug discovery library needs to account for not only quantity, but also quality of compounds. DEL technology has never had a problem with the former, as nearly any number of compounds can be simultaneously screened once in the library, but has historically struggled with the latter. The majority of DEL libraries are built with a number of cycles of split-and-pool chemistry, which can quickly lead to the generation of compounds well outside the bounds of traditionally “drug-like” chemical space.
DyNAbind’s fragment-based library, however, has been designed to ensure that any identified hits will be valid medicinal chemistry starting points. All fragments used in the library have been pre-filtered against an in-house list of hundreds of substructures to remove promiscuous, toxic or reactive substructures. Final selections for compound inclusion were made with the help of chemical fingerprinting to maximize compound diversity. The average Tanimoto similarity for any two of the fragments in DyNAbind’s collection is less than 0.2.
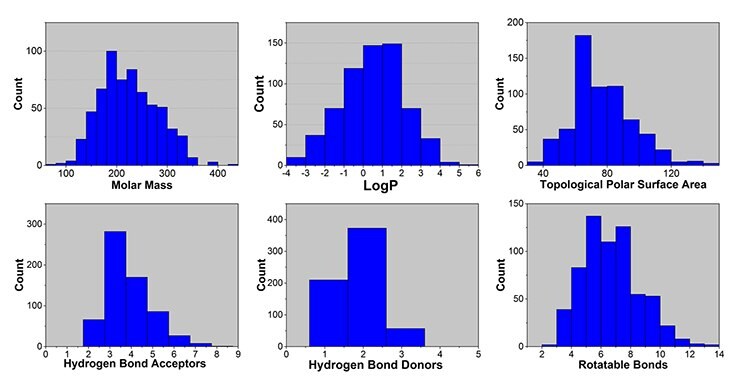
As seen in the histograms below, the library members’ physicochemical properties were selected to allow ample flexibility in downstream medicinal chemistry work.
Advantages of DNA-Encoded Library Technology
- Unprecedented access to large library drug discovery
- Dynamic Library technology offers more reliable results than classical DEL approaches
- Library members selected for MedChem relevance
- High chemical diversity, suitable for a number of targets
Materials
Dynamic DNA-Encoded Libraries
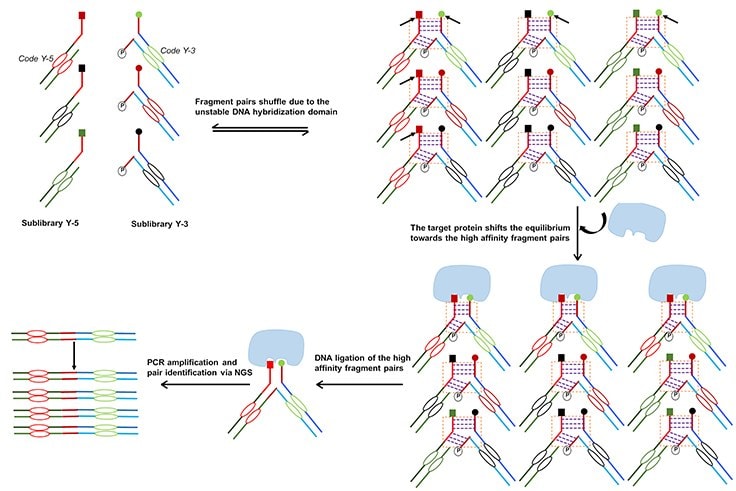
Figure 1. Dynamic DNA-Encoded Libraries.Two sublibraries (5’ and 3’ functionalized) of DNA-Encoded compounds are mixed together where they randomly assemble into fragment pairs. They constantly reshuffle in solution due to the transiently-tuned DNA interactions. Upon introduction of the target protein, the thermodynamic equilibrium is shifted, as high-affinity fragment-pair binders are then stabilized by binding to the target. At the end of the selection process, the secondary encoding strands are ligated together and PCR amplified to provide information on the pairing of binders.
For compounds that are further along in development, we also offer the 10 Million Compound DNA-Encoded Library (DYNA002). This library allows users to find hits composed of three fragments instead of just single or paired fragments. If the extended DEL Data (DELDATA) package is purchased, the researcher is given access to all data revealed by the 10 Million Compound library instead of just the first 50 hits. This allows researchers to utilize AI and machine learning to learn even more information on their targeted compound.
Webinars
- A New Screening Approach for Lead Discovery: DNA-Encoded Library Technology
This webinar covers a new approach to lead discovery and screening compounds, presented in partnership with DyNabind. Watch to learn more about our DNA-encoded library technology.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?