Detection of Campylobacter spp. and Enterobacter Sakazakii in Food with a New Test Kit System
Fabienke, Scanbec GmbH; Siegrist, Product Manager Microbiology, Sigma-Aldrich ivo.siegrist@sial.com
Background
Development of novel methods for a rapid, sensitive and reliable detection and quantification of microorganisms and pathogens in food, beverages and water is receiving increasing attention. The sandwich hybridisation method used in the HybriScan® Test System is a suitable alternative for a sensitive and reliable detection and identification of microorganisms.
The HybriScan method is nearly independent of influences of sample matrices, and is able to distinguish between live and dead cells. Furthermore, the detection of non-culturable microbes is possible.
The HybriScan method is based on the detection of hybridisation events between two specific oligonucleotide probes and target nucleic acids. The capture probe is used to immobilise the target sequence on a solid support and the detection probe is labeled with a detectable marker (Figure 1). Sandwich hybridisation is relatively sensitive and can be performed with crude biological samples1. Sandwich hybridisation assays from crude cell samples or in connection to PCR have been extensively used in clinical diagnostics for detection of nucleic acids from bacteria2, 3, 4 and viruses5. The sandwich hybridisation method is ideal for identification of specific rRNAs in bacterial cells and yeasts. The sensitivity of this RNA-based assay benefits from the typically high number of ribosomes in each cell. Compared to only a few copies of genomic DNA a single cell contains several thousand copies of rRNA. Although a direct detection of the ribosomal RNA does not match the sensitivity of a PCRbased DNA assay, it offers advantages like quantification, live/ dead-discrimination, no additional amplification steps and simple assay protocols with standard laboratory equipment.

Figure 1.Principle of the HybriScan sandwich hybridisation assay
HybriScanD Campylobacter: Detection of Campylobacter spp. in food
Bacteria of the species Campylobacter are zoonotic pathogens which annually infect 1 % of Western Europe’s population. Some Campylobacter species are known to infect animals, especially infections of the reproductive tract.
Campylobacter are ubiquitous and often found in domestic animals. In this way, they are present frequently in the environment and on many raw foods of plant and animal origin. A very high concentration of Campylobacter can be found on raw poultry meat6.
According to authorities in Germany, human Campylobacteriosis is the most common reported disease caused by bacteria besides Salmonellosis. In 2004, around 55,000 and in 2005, 60,000 Campylobacter infections were observed7.
Therefore a rapid and reliable detection of Campylobacter is required to ensure microbiological safety and quality. In contrast to HybriScan, classical cultural methods for the detection of Campylobacter spp. are time consuming and well-trained laboratory personnel is required for each type of bacteria.
HybriScanD Campylobacter is a rapid molecular test system for the detection of bacteria of the genus Campylobacter in different food matrices, including the detection of the most relevant species C. jejunii (Figure 2), C. coli, C. lari and C. upsaliensis.
Figure 2.Campylobacter jejunii colonies grown on CCDA agar
HybriScanD Campylobacter enables a reliable and comprehensive control of suspicious results in the context of classical microbial diagnostics but makes detection more rapid, with results available after 48 hours. An overview of the validation results of HybriScan- Campylobacter spp. are presented in Figure 3. 108 food samples were analysed with HybriScanD Campylobacter and compared with a cultivation-based method according to § 64 LFGB. In total, five different food categories were tested. The results of the validation lead to a relative accuracy of 95.2 %, a relative specificity of 97.5 %, and relative sensitivity of 93 %, respectively.
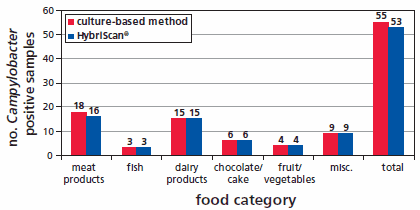
Figure 3.Overview of the validation results of HybriScan®D Campylobacter 108 food samples were analysed with HybriScanD Campylobacter and compared to a cultivation-based method according to § 64 LFGB. Numbers on the bars represent the number of analysed food samples in each food category. The validation was performed according to ISO 16140:2003 (ASU L00.00-22).
HybriScanD Enterobacter sakazakii: Detection of Enterobacter sakazakii in food
Enterobacter sakazakii is ubiquitous and frequently found on vegetables, meat and dairy products, and especially in baby food. Consumption of contaminated powdered infant formula milk (IFM) can result in life-threatening neonatal infections caused by the pathogen. Taxonomic studies have determined that E. sakazakii comprises a high genetic heterogeneity and should be reclassified as a novel genus, “Cronobacter”8 (Figure 4).

Figure 4.Enterobacter sakazakii colonies grown on M1 agar
Accurate methods are required for the rapid detection and identification of Enterobacter sakazakii, since even low cell numbers have been reported to cause a disease. HybriScanD E. sakazakii is a rapid molecular test system for detection of bacteria of the genus E. sakazakii in food, especially in dried infant formula milk and its production environment. Figure 5 shows the specificity of HybriScanD E. sakazakii. Different cell amounts and related Enterobacteriaceae were tested within a validation study. No signals were observed using 2.3 x 108 Enterobacter cloacae cells or 7 x 108 Citrobacter freundii cells per assay, whereas clear specific signals were detectable using 2.6 x 103, 1.3 x 104, and 2.6 x 104 cells of E. sakazakii, respectively. These results demonstrate that the HybriScan system is highly specific for Enterobacter sakazakii.
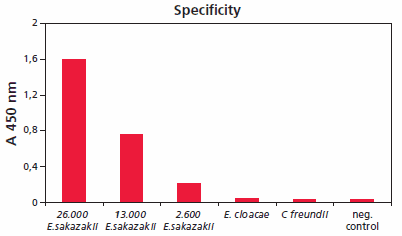
Figure 5.Specificity of HybriScanD E. sakazakii. Different cell numbers of E. sakazakii and related Enterobacteriaceae like E. cloacae and Citrobacter freundii were tested. Measurement data for HybriScan analyses represent absorption at 450 nm.
A validation study of HybriScanD E. sakazakii was performed using two different enrichment procedures: (1) single-step enrichment for 24 –26 hours at 37 °C in ESSB broth (AES Chemunex) and (2) twostep enrichment starting with a pre-enrichment for 18 –20 hours at 37 °C in buffered peptone water and followed by a selective enrichment for 24 –26 hours at 45 °C in mLST selective broth. The results of the above-mentioned validation study are presented in Table 2.
130 samples of powdered infant formula milk were analysed with HybriScanD E. sakazakii and compared to two cultivation-based methods according to ISO/PFR TS 22964. In total, six products from different manufacturers were tested. Results of the validation study lead to a relative accuracy of 95 %, a relative specificity of 92.9 % and relative sensitivity of 95.7 %, respectively.
HybriScanD E. sakazakii enables a reliable and comprehensive control of suspicious products in the context of classical diagnostic but makes detection more rapid with results available after 48 hours.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?