Using Microarray Technology to Select Housekeeping Genes in Chinese Hamster Ovary Cells
Abstract
In the present study, we have identified species-specific housekeeping genes (HKGs) for Chinese HamsterOvary (CHO) cells using data from microarray gene expression profiling. HKGs suitable for quantitativeRT-PCR normalization should display relatively stable expression levels across experimental conditions. Weanalyzed transcription profiles of several IgG-producing recombinant CHO cell lines under numerous culture conditions using a custom CHO DNA microarray platform and calculated relative expression variability from124 microarrays. We selected a novel panel of candidate HKGs based on their low variability in expression from the microarray data. Compared to three traditional HKGs (Gapdh, Actb and B2m), the majority of genes on this panel demonstrated lower or equal variability. Each candidate HKG was then validated using qRT-PCR. Final selection of CHO-specific HKGs include Actr5, Eif3i, Hirip3, Pabpn1, Vezt, Cog1 and Yaf2. The results reported here provide a useful tool for gene expression analyses in CHO cells, a critical expression platformused in biotherapeutics.
Materials and Methods
Cell Culture and RNA Isolation
The cell culture experiments used in the present study involved 25 different experimental growth conditions including two parental and four IgG-producing CHO cell lines (CHO K1 lineage). All cell lines used were cultured in serum-free, animal component free suspension media. Our dataset represented batch and fed-batch cultures in 125-ml shake flasks, TPP bioreactor tubes and 6-well static cultures, with numerous chemically-defined or plant hydrolysate containing media and treatments such as sodium butyrate. All experiments were conducted in biological duplicates. Parental and recombinant cell lines were all included in the analyses. Cells were collected during the early-and mid-logarithmic as well as stationary phases for RNA isolation. Total RNA isolation was performed with RNeasy Mini Kit (Qiagen, Valencia, CA). RNA concentrations were measured using Nanodrop 1100 (Nanodrop Technologies, Wilmington, DE). RNA integrity was analyzed with Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Microarray Design, Hybridization and Expression Data Analysis
An internally developed CHO DNA sequence database containing over 60,000 total sequences and 9,000 annotated contigs was used to design the 4x 44K custom microarray platform (Agilent Technologies, SantaClara, CA). The average probe length is 60-bp, with a melting temperature (Tm) of 70 °C and a GC content of 45%. Each sequence has two probes designed to hybridize with different regions. In the instance that known genes were not represented in our sequence database, mouse orthologous gene sequences were used for probe design. This array platform was used in all 124 microarray studies.
A common RNA reference pool was created from an assortment of CHO lines and conditions in order todirectly compare across experiments. Dye-swap technical replicates were conducted as following: RNAsamples were labeled with Cy5 or Cy3 and hybridized against the common reference pool labeled with Cy3or Cy5, respectively.
Sample labeling, amplification and hybridization were performed with the 2-Color Low RNA Input LinearAmplification Kit (Agilent Technologies). Slides were washed in acetonitrile (Sigma-Aldrich, St. Louis, MO) and treated with Stabilization and Drying Solution (Agilent Technologies) according to manufacturer’sprotocol. Agilent Feature Extraction software 9.5 (FE 9.5) was used to perform dye normalization and QC statistics based on manufacturer’s protocols.
Statistical Analysis
The relative expression levels of each probe were normalized using the median fluorescence intensity of thereference pool in each dye-swap set. CHO-specific HKG candidates were selected using the following criteria. The coefficient of variation (% CV) was calculated using the expression value of each probe normalized with the median intensity of the reference pool. Each gene must have at least one microarray probe with a % CVbelow 29.21 % (25% percentile of all probes analyzed, (Figure 1).
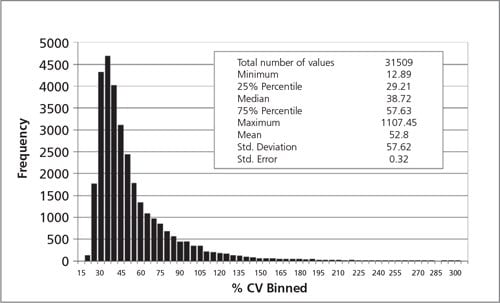
Figure 1.Depicts the distribution of the % CV values from the 31509 probes on 124 microarrays used for thepresent analysis. Quality control and background estimation probes on the microarray were excluded. The 25%percentile value is 29.21%, which was used for the primary cutoff for CHO HKG selection. The data points (n =232) with % CV greater than 300% were not shown on this histogram.
Quantitative RT-PCR Validation
Primers were designed against sequences from the internal CHO DNA sequence database using Primer3 software (http://fokker.wi.mit.edu/primer3/input.htm). The primers were 18–25 base pairs (bp) with Tm of64–65 °C. The amplicons were between 156–249 bp (Table 2). All amplification efficiencies were calculated based on at least 4 dilution points and within a range of 90%–112%.A minimum of ten experimental conditions were randomly selected for qRT-PCR validation, with biological duplicates from the microarray experiments pooled for RT reactions. qRT-PCR was performed on a MX3000 Pthermocycler (Stratagene, La Jolla, CA) in triplicate for each experimental condition, with threshold cyclevalues (Ct) averaged from the triplicates. Reactions were run with SYBR Green Jumpstart™ Taq ReadyMix™ (Sigma-Aldrich, S4438) mixed with 1 mL of cDNA prepared from 25 ng/mL RNA and primers at 500 nM in areaction volume of 20 mL.
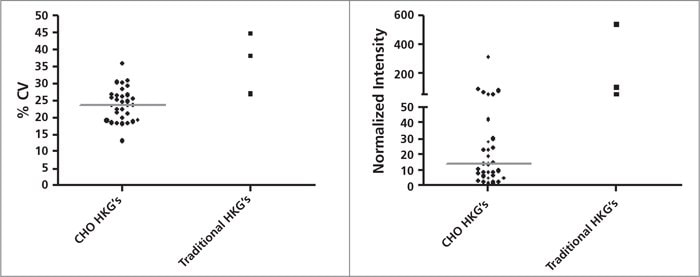
Figure 2.Left panel: % CV distribution of 31 probes of the 12 CHO HKG candidates and three traditional HKGs(Gapdh, Actb and B2m). Right panel: Relative expression level distribution of the 12 CHO HKG candidates and three traditional HKGs.Each dot in the scatter dot plots represents one microarray probe. Horizontal line represents the median. Averageof all the probes of each gene is depicted for the traditional HKGs.
Conclusions
- We are the first to report a novel panel of species-specific housekeeping genes (HKG) for CHO cells. These genes were selected based on expression stability from data collected from more than 25 experimental conditions in various CHO cell lines using our custom microarray platform.
- In qRT-PCR validation, the expression stability for our HKG panel is superior or comparable with commonly
used HKG’s such as Actb, B2M, and Gapdh. - Our HKG panel includes several low expression genes that may be useful for normalization of low abundance transcripts. These HKG’s can be used in conjunction with commonly used HKG’s for more accurate data normalization.
Acknowledgements
Carol Kreader, Brian Ward, Christoph Bausch, Yang Liu – Sigma Aldrich Biotech
Materials
如要继续阅读,请登录或创建帐户。
暂无帐户?