Absolute Quantification of Serum Proteome
Plasma proteomics holds great promise for the future of biomarker discovery, as well as in vitro diagnostics. Although plasma is readily accessible for analysis, the study of the plasma proteome is fundamentally limited by its vast dynamic range (10 orders of magnitude). Low-abundance proteins, carrying great diagnostic potential, are often obscured by the presence of high-abundance serum proteins. In order to address this problem, we have developed the ProteoPrep® 20, an antibody-based resin capable of depleting 97–98% of the total protein from plasma. Depletion of these high-abundance proteins allows for visualization of proteins co-eluting with, and masked by, the high-abundance proteins and peptides, using LC-MS methods. Absolute Quantification (Protein-AQUA™) is a targeted quantitative proteomics technique that exhibits robust efficacy and is being increasingly utilized for a wide variety of quantitative proteomics studies. By applying the ProteoPrep® 20 for depletion of the most abundant proteins, we have enabled the identification and absolute quantification of numerous lower- abundance proteins from human serum. We believe this may potentially provide a multiplexed targeted approach to the discovery of proteins holding biological or diagnostic significance.
Introduction
The study of the human plasma proteome is an area of great interest, especially for the pharmaceutical potential of identifying disease biomarkers. Many proteins of pharmaceutical interest appear at low concentrations in the plasma and are, therefore, difficult to detect. Identification of potential biomarkers is especially difficult due to the presence of higher- abundance proteins. Depletion of these high-abundance proteins allows for visualization of proteins co-eluting with, and masked by, the high-abundance proteins and peptides, using LC-MS methods. By removing the high-abundance proteins, higher loads of lower-abundance plasma proteins can then be separated on reverse phase columns, allowing detection and quantification of low copy number proteins. The ProteoPrep® 20 Plasma Immunodepletion Kit has been developed for removal of 20 high-abundance proteins from 8 mL of plasma. Depletion of these 20 high-abundance proteins removes greater than 97% of the proteins in plasma and permits analysis of 20- to 50-fold more of each individual protein for improved visualization of lower copy number proteins. By applying the ProteoPrep® 20 for depletion of the 20 most abundant proteins, we have enabled the identification and absolute quantification of numerous lower-abundance proteins from human serum.
Plasma Facts
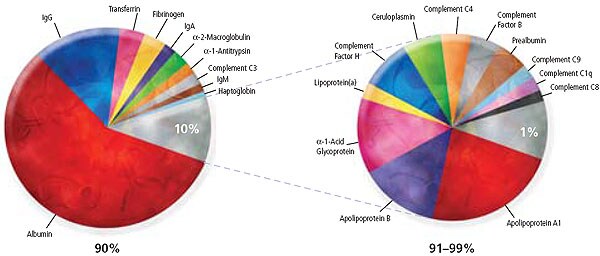
Figure 1.AQUA™ Depletion plasma facts
- The 10 most abundant proteins represent approximately 90% of the total protein mass in human plasma.
- The 22 most abundant proteins are said to represent approximately 99% of the total protein mass in human plasma.
- The ProteoPrep® 20 plasma immunodepletion column removes the 20 high-abundance plasma/serum proteins listed below. These 20 proteins represent approximately 97% of the total human plasma protein mass.
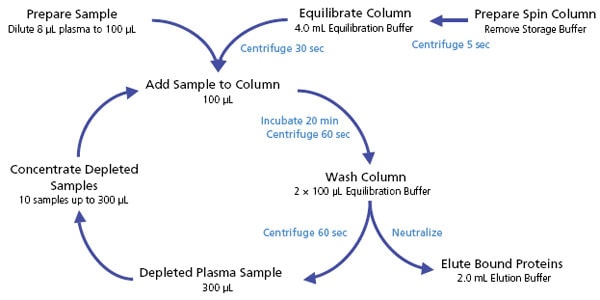
Figure 2.Plasma Protein Depletion Workflow
Methods
High-Abundance Protein Depletion
Twenty high-abundance proteins were depleted from plasma using the ProteoPrep® 20 Immunodepletion Kit (PROT20). A large 3.7-mL prototype spin column capable of depleting 100 mL of human plasma was used. The protein concentration of whole and depleted plasma was determined using Bradford assay (B6916), with BSA as the standard.
Tryptic Digestion
Samples of both whole and depleted plasma were prepared in 100 mg aliquots. Each sample was diluted to 0.25 mg/mL in ammonium bicarbonate buffer. Acetonitrile was added to a final concentration of 9% (v/v), and each sample was reduced and alkylated using the ProteoPrep® Reduction/Alkylation Kit (PROTRA). Proteomics Grade Trypsin (T6567) was added at a concentration of 1% (w/w) and the sample incubated for 3 hours at 37 °C. An additional aliquot of trypsin at 1% (w/w) was added and then the sample further incubated at 37 °C overnight. The digests were dried to completion in a SpeedVac™.
LC-MS/MS Analysis
An injection of a tryptically digested ProteoPrep® 20 depleted plasma sample (prepared as described in Methods) was made using an Agilent capillary 1100 HPLC. The reverse phase separation was performed on a 15 cm × 2.1 mm Discovery C18 column, using a 3-hour gradient. The mobile phase consisted of formic acidified water and acetonitrile. The separation system was coupled to a Thermo Finnigan LTQ linear ion trap mass spectrometer set to perform tandem MS on the ten most abundant ions in each full scan spectrum. After obtaining tandem MS results on an ion of interest, it was added to an exclusion list to facilitate the interrogation of lower-abundance ions. An LC-SRM method was developed corresponding to the protein targets albumin (ideotypic peptide = FQNALLVR) and gelsolin (ideotypic peptide =GASQAGAPQGR), as outlined in the Results section.
AQUA™ Analysis
An AQUA™ Peptide stock solution was prepared by dissolving isotopically labeled versions of the two target peptides (FQNALLVR* and GASQAGAPQGR*, respectively) in 0.1% TFA to a final concentration of 62.5 fmol/mL. Each digest sample (whole and depleted plasma) was dissolved in 20 mL of the AQUA™ Peptide stock solution. Samples were analyzed using an Agilent capillary 1100 HPLC coupled to a Thermo Finnigan LTQ linear ion trap mass spectrometer. Using an LC-SRM method, the absolute quantity of each peptide (and corresponding protein) was determined.
Results
![Overview of the Protein-AQUA™ Method. This method was developed by Dr. Steve Gygi and colleagues at Harvard Medical School [Stemmann O., Zou H., Gerber S. A., Gygi S. P., Kirschner M. W.; Dual inhibition of sister chromatid separation at metaphase, Cell 2001, Dec 14, 107: 715-726]. Limited use of this method is permitted under a licensing arrangement with Harvard Medical School. Overview of the Protein-AQUA™ Method](/deepweb/assets/sigmaaldrich/marketing/global/images/technical-documents/articles/protein-biology/protein-mass-spectrometry/aqua-depletion-method.gif)
Figure 3. Overview of the Protein-AQUA™ Method. This method was developed by Dr. Steve Gygi and colleagues at Harvard Medical School [Stemmann O., Zou H., Gerber S. A., Gygi S. P., Kirschner M. W.; Dual inhibition of sister chromatid separation at metaphase, Cell 2001, Dec 14, 107: 715-726]. Limited use of this method is permitted under a licensing arrangement with Harvard Medical School.
Select an optimal tryptic peptide and stable isotope amino acid from the sequence of your protein of interest.
Order synthetic AQUA™ Peptide from.
Optimize LC-MS/MS separation protocol for quantitation.
Extract protein from biological samples and add known quantity of AQUA™ peptide.
Digest.
Analyze by LC-MS/MS or MALDI to quantitate protein of interest.
Figure 4. Development of a Protein-AQUA™ LC-SRM for Absolute Protein Quantification. An injection of a tryptically digested ProteoPrep® 20 depleted plasma sample (prepared as described in Methods) was made using an Agilent capillary 1100 HPLC coupled to a Thermo Finnigan linear ion trap mass spectrometer. The base peak chromatogram (Figure 2A) shows a complex mixture of species that may or may not have been chromatographically resolved. By harnessing the power of tandem MS, more than 100 proteins were positively identifi ed, with confidence, from the depleted plasma sample. Two of these proteins (albumin and gelsolin) were chosen for further study. The full scan MS shown in Figure 2B illustrates the high abundance of the 481 Da doubly charged ion from one of the albumin peptides identified. This sequence is then confirmed in Figure 2C with an abundance of y- and b-ions. This experimental approach lays the groundwork for choosing an appropriate AQUA™ peptide in addition to providing the necessary information for designing the SRM experiment. As shown in Figures 2B and 2C, the doubly charged parent ion of this specific albumin peptide was selected for fragmentation and the two most abundant y-ions present in the tandem MS were selected for detection.
Using the ProteoPrep® 20 depletion technology, we have enabled the identification and absolute quantification of these proteins. Data sets were searched using the SEQUEST algorithm in Bioworks 3.2. Filtration of protein hits consisted of XCORR values >1.9, 2.2, and 3.75 for singly, doubly, and triply charged ions, as well as delta CN > 0.1 and Rsp > 4.
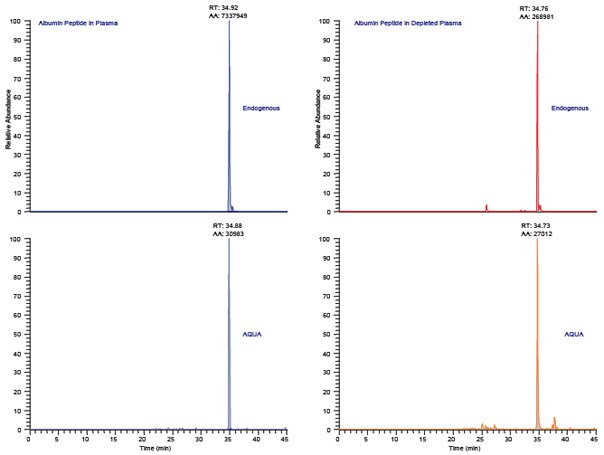
Figure 3.Protein-AQUA™ Analysis of Albumin in Whole Plasma and ProteoPrep® 20 Depleted Samples. Selected reaction monitoring (SRM) specifically targets MS detection of the selected parent that produces specific daughter ions. Figures 3A (whole plasma) and 3C (depleted) depict the endogenous albumin peptide, FQNALLVR, as the doubly charged ions fragment to produce two abundant y-ions. These same y-ions are monitored in the AQUA™ peptide standard shown in Figures 3B (whole plasma) and 3D (depleted); however the absolute mass-to-charge of both parent and daughter ions in the AQUA spike are of higher mass due to the incorporation of a stable isotope amino acid. Quantitation is performed by integrating the co-eluting peak areas shown here at 34.8 minutes. Protein-AQUA™ analysis of the samples indicates 99.8% depletion of albumin using the ProteoPrep® 20 Plasma Immunodepletion Kit, which is in excellent concordance with previously obtained ELISA data.
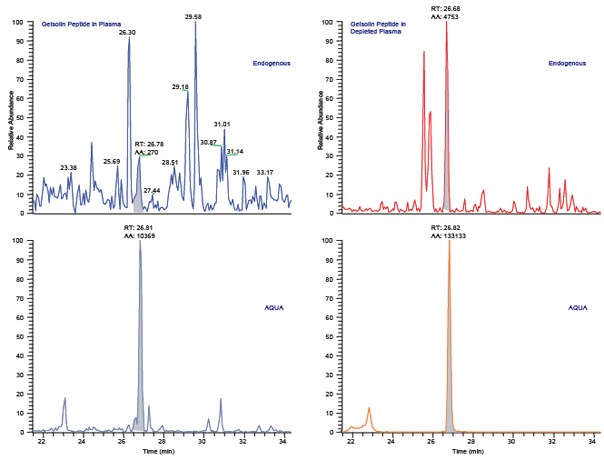
Figure 4. Protein-AQUA™ Analysis of Gelsolin in Whole Plasma and ProteoPrep® 20 Depleted Samples. SRM Analysis of the endogenous gelsolin peptide, GASQAGAPQGR, results in a complex chromatogram in whole plasma (Figure 4A) with poor resolution and low signal-to-noise. When analyzed in depleted plasma (Figure 4C), the endogenous gelsolin peptide is easily separated from interfering species and its concentration readily calculated by comparison to the AQUA™ peptide internal standard (Figures 4B and 4D).
Conclusions
Plasma proteomics holds great promise for the future of biomarker discovery, as well as in vitro diagnostics. Although plasma is readily accessible for analysis, the study of the plasma proteome is fundamentally limited by its vast dynamic range (10 orders of magnitude). Low-abundance proteins, carrying great diagnostic potential, are often obscured by the presence of high abundance serum proteins. By applying the ProteoPrep® 20 for depletion of the most abundant proteins, we have enabled the identification and absolute quantification of numerous lower-abundance proteins from human serum. We believe this combination of technologies may potentially provide a multiplexed targeted approach to the discovery of proteins holding biological or diagnostic significance.
References
如要继续阅读,请登录或创建帐户。
暂无帐户?