Universal Proteomics Standards
The era of high-throughput proteomics has recently blossomed due in large part to advances in the methods by which proteins and proteomes are analyzed. Improved fractionation techniques, combined with advances in mass spectrometry, have decreased concerns of sample complexity, and directed more focus towards high-throughput techniques. However, the reliability of the generated data can be questionable, oftentimes without the knowledge of the data recipient.1 Misidentification of proteins, largely due to the rate of false positives generated by many proteome analyses, has led many scientists to express concern over the accuracy of published proteomics data within the literature. As a result, journals have taken steps towards standardizing proper data collection, analysis, interpretation and reporting.2 However, the high rate of false positives and question of data validity still remains.
The need for the determination of appropriate confidence limits and thresholds for large data sets in proteomics experiments was first demonstrated several years ago through the creation and analysis of artificial mixtures of proteins and peptides. Eugene Kolker’s group created a set of standards using commercially available components, and then used the mixture to verify their data and experimental workflows.3 His group and others have since produced several versions of this mixture and continue testing the mixture against typical proteomic workflows. Their results have profoundly demonstrated the need for the regular, systematic testing of known samples or standards to minimize the false discovery rate of both proteins and peptides. Additionally, the use of standards has been shown to validly address issues in determining instrument/technique reliabilities, setting appropriate confidence limits with large datasets, and the cross-comparison of data sets among collaborating labs. The need for a commercially available, consistently produced protein mixture to serve as a standard was clear.
In early 2005, Merck entered into collaboration with a subgroup of the ABRF (Association of Biomolecular Resource Facilities), the sPRG (Proteomics Standards Research Group) to produce a well-defined complex protein standard for use in the proteomics community. The mission of sPRG is to “promote and support the development and use of standards in proteomics,” and they were interested in conducting a study which was designed to gauge the pulse of the proteomics community and determine how well (or poorly) the participating labs could identify the components of a known complex mixture of proteins. Following several trial batches, a mixture of 49 human proteins was produced and tested by 74 participating labs. The results of this study were presented by Jeff Kowalak at the ABRF annual meeting in 2006, with some surprising results.4,5 A significant number of the respondents (66%) were able to identify at least 30 out of the 49 proteins, with only 8% identifying <10 of the proteins. While the majority of the respondents used similar analysis/identification methods, the sPRG found that no one approach was particularly better than any other. The consortium reconciles this ambiguity by concluding that the success in protein identification was more likely experience- or user-dependent rather than method- or platform-dependent. The study also noted that good results are possible in laboratories that do not have the most sophisticated instruments, but instead spend time optimizing experimental variables. Further results of the study can be found on the ABRF website.6
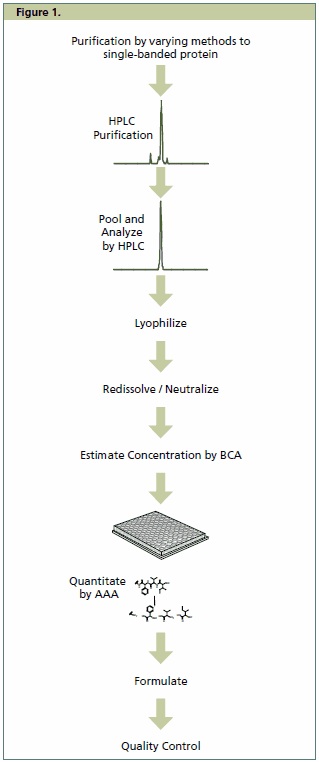
Figure 1.Production of the Universal Proteomics Standards line follows a streamlined process.
This large collaborative effort helped to not only identify the important characteristics of a proteomics standard, but also led to the development of a number of criteria which were deemed important in its production. Using the sPRG study results as a foundation, and through a number of iterative improvements, Merck released the first commercially available proteomics standard in the fall of 2006, Universal Proteomics Standard (UPS1). The product consists of 48 HPLC purified human proteins in equimolar amounts. Streamlining of the purification and production process constituted a considerable challenge for production of the standard. A summary of the process used to produce the protein components of the Universal Proteomics Standard family is shown in Figure 1. Following a variety of purification methods to obtain proteins which are single-banded by SDS-PAGE analysis, all proteins in the standard are purified further by reverse phase HPLC. Following lyophilization and reconstitution of the purified material in water, a microBCA (QPBCA) assay is used to estimate protein concentration, and HPLC analysis is performed to determine protein purity. Final quantitation is by amino acid analysis (AAA), which is considered by many as the most accurate form of protein quantitation. For UPS1, the proteins are then formulated in equimolar amounts (5 pmoles each per vial) of 48 human proteins. As of this writing, the product is the most complex proteomics standard available, and thus the truest mimic of the proteome.
Merck has endeavored to ensure that the Universal Proteomics Standard line of products are produced to meet a rigid set of criteria for purity, accurate quantitation, reliability and reproducibility (Table 1). The high production standards which have been set forth dictate the need for these standards to be produced in the same fashion every time. The maintenance of collaborative efforts ensure that the Universal Proteomics Standards continue to meet the needs of the scientific community, a fact which is already being observed. For example, Christoph Turck’s group utilized the mixture as a means of developing new separation and search parameters leading to higher hit confidence and decreased number of false positives.7 In a similar vein, David Tabb’s group demonstrated the use of the UPS1 standard as a testing platform for their newly developed scorer, MyriMatch.8 Rune Matthieson’s lab utilized UPS1 standard as a negative control for glycophosphatidylinositol proteins, allowing them to verify that their search parameters were not identifying false postives.9
Although valuable in many respects, an equimolar protein standard does fail to account for the broad dynamic range found in most proteomics samples. For example, plasma is thought to contain ten orders of magnitude of protein levels. However, mass spectrometers are not currently capable of reliably analyzing across such a broad dynamic range. Therefore, Merck has recently introduced the second addition to its standards line; the Dynamic Range Standard (UPS2), a product which is designed to push the envelope of testing by mass spectrometry. This standard uses the same 48 proteins as offered in UPS1, however, the proteins have been divided across six concentration ranges spanning five orders of magnitude (50 pmol – 0.5 fmol, per vial). As shown in Table 2, the components in each level were specifically chosen to present diverse molecular weights, isoelectric points, and hydrophobicities. The dynamic range standard has been designed to appear much like a real proteomics sample, allowing researchers to not only tune their instrumentation, but also their separation methods, prior to analysis of their real sample.
Testing and validation of the value of UPS2 was accomplished through continued collaboration with a large number of scientists in the proteomics community. The ability to detect proteins across a wide range of concentrations using mass spectrometry relies largely upon separation techniques at both the protein and peptide level, and is, to say the least, extremely difficult. Results from a number of colleagues who have tested the dynamic range standard have illustrated the need for advanced fractionation to observe even three or four orders of magnitude. The ability to confidently identify proteins across the five orders of magnitude, as presented in the Dynamic Range Standard, requires continual improvement and refinement to both instrumentation and separation methods. The use of standards in this iterative process can greatly facilitate the ability to push the envelope further, and potentially reach previously unobserved low level proteins and biomarkers. The use of standards provides insight into the effectiveness of everyday tasks prior to, and during sample processing. As the data sets being analyzed continue to grow in size and complexity, and the analysis techniques continue to become more reliable and sensitive, it still remains important to maintain appropriate confidence limits, as well as understand the potential for, and occurrence rate of, false positives. The use of defined protein mixtures remains invaluable in this regard. However, as a learning tool, standards are also invaluable: allowing the end-user the ability to vary and tweak their protocols and instruments to optimize the performance of their workflows. As new techniques and instrumentation continue to evolve, the need for standards will also continue to grow.
Related Products
References
如要继续阅读,请登录或创建帐户。
暂无帐户?