Purification of Membrane Proteins
Membrane proteins are usually purified as protein-lipid-detergent complexes. The solubility of the complexes in an aqueous environment allow the application of essentially the same separation techniques as used for water-soluble proteins. The main difference is that the purification of membrane proteins is carried out with detergent present in all solutions. This is necessary because protein-detergent complexes are dynamic and would immediately lose detergent molecules in the absence of free detergent. Detergent concentrations should be above the CMC but can be kept about 10 times lower than what was used during solubilization (typically in the 0.1% range).
High detergent concentrations can reduce the stability of the protein. However, detergent concentrations need to be high during solubilization if the concentration of membrane components is high. Once solubilization is completed, detergent concentrations can be reduced. Detergents are often expensive, and it is also useful to limit consumption for cost reasons.
Over-purification can lead to the removal of essential lipids from the protein-lipid detergent complex with concomitant loss of protein activity.
Membrane protein stability can often be improved by having 5% glycerol in all buffers throughout the purification.
Purification of histidine-tagged membrane proteins
Histidine-tagged proteins have affinity for Ni2+ and several other metal ions that can be immobilized on chromatographic media using chelating ligands. Consequently, a protein containing a histidine tag will be selectively bound to metal-ion-charged media such as Ni Sepharose High Performance (HP) and Ni Sepharose 6 Fast Flow (FF) while most other cellular proteins will not bind or bind weakly. Elution is achieved by increasing the concentration of imidazole. This chromatographic technique is often termed immobilized metal ion affinity chromatography (IMAC).
Water-soluble, (histidine)6-tagged proteins are usually straightforward to purify following standard protocols. Histidine-tagged membrane proteins are sometimes more problematic, and weak binding to IMAC media is often reported. It has been speculated that this is due to restricted accessibility of the histidine-tag for the IMAC ligand due to the binding of detergent to the protein. To address the issue, longer histidine tags are routinely used for overexpression of membrane proteins. Also, the presence of a linker, such as green fluorescent protein (GFP), between the histidine tag and the target membrane protein has been suggested to improve binding to IMAC media.
Historically, batch-wise purification has often been employed for the purification of histidine-tagged membrane proteins. Batch-wise purification involves mixing the sample with the chromatography media in an open vessel for a designated time, often overnight. The suspension is then packed into a column for washing and elution of the bound protein. Batch-wise purification can sometimes improve yields since adsorption times are longer than for column separations. On the other hand, since a batch-wise procedure is longer it also leaves the protein more exposed to proteolytic degradation or inactivation and may thus compromise the quality of the purified protein.
Purification of histidine-tagged membrane proteins can also be performed using column-based methods.
Depending on the level of purity required for the final application, additional purification steps can be performed after IMAC. For this purpose, gel filtration is possibly the most common, and has the advantage that optimization is usually not needed. Anion exchange chromatography can also be used and is often included between IMAC and gel filtration steps.
Purification of Histidine-tagged Membrane Protein from the Solubilized, Isolated Membrane Fraction Material
Material
Column: HisTrap HP, 1 mL
Binding buffer: PBS, 20 mM imidazole, 0.1 to 1 % detergent (e.g., DDM), pH 7.4
Elution buffer: PBS, 0.5 M imidazole, 0.1 to 1% detergent (e.g., DDM), pH 7.4
Sample preparation
To 5 mL of solubilized membrane protein, add 0.20 mL of elution buffer to give a final imidazole concentration of 20 mM.
Purification
This purification procedure should be performed at 4 °C.
- Fill the syringe or pump tubing with distilled water. Remove the stopper and connect the column to the syringe (use the connector supplied), laboratory pump, or chromatography system “drop to drop” to avoid introducing air into the system.
- Remove the snap-off end at the column outlet.
- Wash out the ethanol with 3 to 5 column volumes (CV) of distilled water.
- Equilibrate the column with 10 CV of binding buffer at a flow rate of 1 mL/min*
- Apply the sample (using a syringe fitted to the Luer connector or by pumping it onto the column). Use a flow rate of 1 mL/min.
- Wash with 10 CV of binding buffer at a flow rate of 1 mL/min.
- Elute with a gradient of 0% to 75% elution buffer in 20 CV at a flow rate of 1 mL/min. When a syringe is used, elute stepwise with successively higher concentrations of imidazole.
- After elution, wash the column with 5 CV 100% elution buffer followed by 5 CV binding buffer.
*One mL/min corresponds to approximately 30 drops/min when using a syringe with a HiTrapTM 1-mL column. When using a larger column, a higher flow rate can be used. See column instructions.
The procedure can be scaled up by connecting two or three columns in series or by using HisTrap HP 5 mL columns.
A relatively low NaCl concentration (e.g., PBS is 150 mM NaCl) is recommended because membrane proteins tend to be less soluble at higher ionic strengths. Higher concentrations (e.g., 300 to 500 mM NaCl) are often recommended for IMAC of water-soluble proteins to reduce ionic interactions of contaminants with the chromatographic medium. For some cases, even lower NaCl concentrations (e.g., < 150 mM) should be applied for a membrane protein. Alternatively, the NaCl concentration can be reduced directly after the IMAC step by desalting the material using a HiTrap Desalting column.
It has been reported that by using gradient elution (with increasing concentrations of imidazole) from an IMAC column, as in the protocol above, protein-lipid-detergent complexes that differ only in lipid content can be separated.
Re-application of the flowthrough material, after step 5 above, to allow the sample to pass through the IMAC column several times can be useful to maximize yield.
The yield can also be increased by decreasing the flow rate during sample loading.
For further purification, ion exchange chromatography and/or gel filtration is often suitable (see “Additional purification steps” in this section).
Figure 1.9 shows the purification of YedZ-TEV-GFP-(His)8 using the protocol above. YedZ is a transporter membrane protein from E. coli that was overexpressed in E. coli as a fusion protein with GFP, a C-terminal (histidine)8 tag and a tobacco etch virus (TEV) protease cleavage sequence.
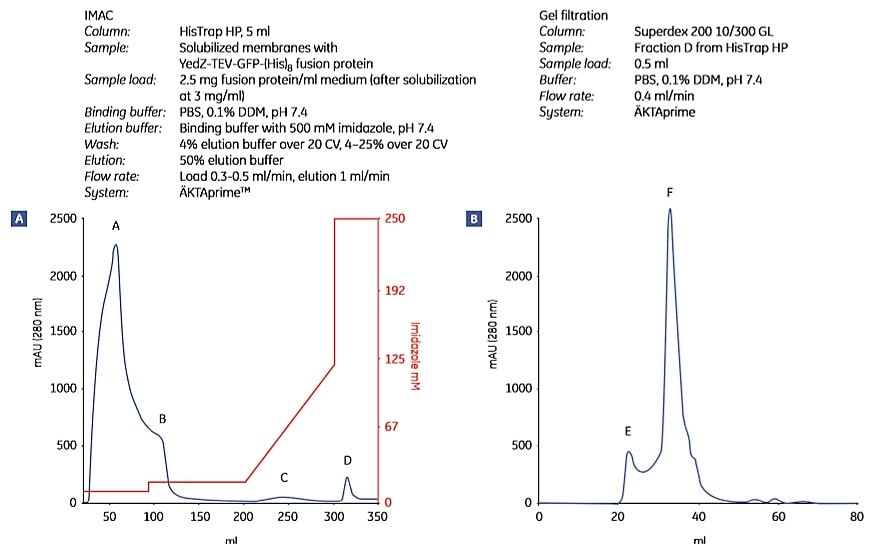
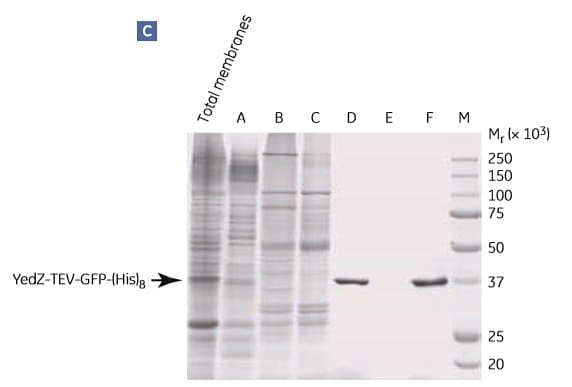
Figure 1.9.Two-step purification of E. coli YedZ-TEV-GFP-(His)8 from solubilized membranes prepared from E. coli cell culture. Fractions from (A) IMAC were further purified by (B) gel filtration. (C) SDS-PAGE analysis of selected fractions shows the purity of the target protein. Fraction D from the HisTrap HP column was essentially homogeneous. M = molecular weight marker. Peak E most likely contained light-scattering detergent-lipid aggregates. Data kindly provided by Dr. David Drew, Center for Biomembrane Research, Stockholm University, Stockholm, Sweden.
Purification of histidine-tagged membrane protein directly from crude, solubilized E. coli lysate
This protocol circumvents the need for membrane preparation and centrifugations. It uses a chromatography column that was designed for the application of unclarified crude cell lysate.
Materials
Column: HisTrap FF crude, 1 mL
Binding buffer: PBS, 40 mM imidazole, 0.1% detergent (e.g., DDM), pH 7.4
Elution buffer: PBS, 1 M imidazole, 0.1% detergent (e.g., DDM), pH 7.4
Sample preparation
Suspend E. coli cell paste by addition of 5 mL PBS with 40 mM imidazole for each gram of paste. Perform cell lysis by lysozyme treatment and sonication (Table 1.2). Add detergent to the unclarified lysate from a concentrated stock (e.g., DDM to a final concentration of 0.8%). Stir on ice for 1.5 hours.
Purification
- Fill the pump tubing or syringe with distilled water. Remove the stopper and connect the column to the chromatography system tubing, syringe (use the provided Luer connector), or laboratory pump “drop to drop” to avoid introducing air into the system.
- Remove the snap-off end at the column outlet.
- Wash out the ethanol with 3 to 5 CV of distilled water.
- Equilibrate the column with 10 CV of binding buffer at a flow rate of 1 mL/min*
- Apply the detergent-treated, unclarified lysate with a pump (0.5 mL/min) or syringe. Loading volumes of unclarified lysate are highly dependent on each specific sample.
*One mL/min corresponds to approximately 30 drops/min when using a syringe with a HiTrap 1-mL column. When using a larger column, a higher flow rate can be used. See column instructions
Continuous, gentle stirring of the sample during sample loading is recommended to prevent sedimentation. Sample loading at 4 °C may increase the viscosity of the sample. An adverse effect of increased sample viscosity is that maximum back pressure for the column is reached at a lower sample volume loading on the column. Large volumes may increase back pressure, making the use of a syringe more difficult.
- Depending on the sample volume (larger sample volumes require larger wash volumes), wash with 10 to 30 CV of binding buffer at a flow rate of 1 mL/min.
- Elute with a gradient of 0% to 12% elution buffer in 10 CV followed by 12% to 100% elution buffer in 5 to10 CV, all at a flow rate of 1 mL/min. When a syringe is used, elute stepwise with successively higher concentrations of imidazole.
- After elution, wash the column with 5 CV elution buffer followed by 5 CV binding buffer.
Figure 1.10 shows the purification of YedZ-TEV-GFP-(His)8 using the protocol above.
For further purification, ion exchange chromatography and gel filtration is often suitable (see “Additional purification steps” in this section).
The procedure can be scaled up by connecting two or three columns in series or by using HisTrap FF crude 5 mL columns.
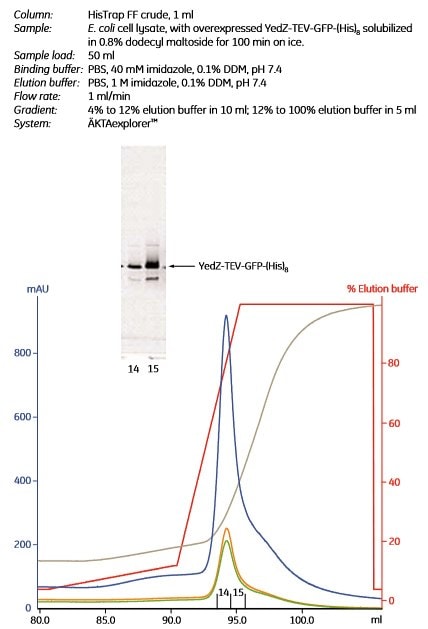
Figure 1.10.Purification of a (histidine)8-tagged membrane protein, YedZ-TEV-GFP-(His)8 directly from crude, solubilized E. coli lysate using HisTrap FF crude, 1 mL. Peak fractions 14 and 15 were analyzed by SDS-PAGE. The gel was scanned to detect the GFP portion of the fusion protein. An arrow indicates the band corresponding to YedZ-TEV-GFP-(His)8. In the chromatogram, blue =A280; orange = A425 (to detect YedZ); green = A485 (to detect GFP); gray = conductivity; red = % elution buffer. The overexpression vector was kindly provided by Dr J.-W. deGier, Centre for Biomembrane Research, Stockholm, Sweden.
Additional purification steps
Further purification of the IMAC-purified, histidine-tagged membrane protein is usually necessary for applications that require highly pure homogeneous material (e.g., for structural characterizations). One or two additional chromatographic steps are usually sufficient. For maximum efficiency, the purification scheme should be designed so that different separation principles are utilized in the different steps.
A highly efficient purification scheme may consist of IMAC (separation according to specific affinity) followed by desalting and ion exchange (separation according to charge differences) and finally by gel filtration (separation according to size differences). Gel filtration is most often used as the last step for the final removal of aggregates and for transfer of the sample to a buffer suitable for further functional and structural studies. The separations are carried out in the presence of detergent above the CMC (typically about 0.1%). Other conditions are the same as for water-soluble proteins. Some suitable columns are listed in Table 1.6. ÄKTAxpress™ chromatography system can be used to automate multistep purifications to produce highly pure proteins with minimum hands-on time.
High-resolution anion exchange can be used for both purification and characterization of the charge homogeneity of purified membrane proteins. In most cases the ionic strength of samples from IMAC must be reduced before application to ion exchange chromatography. Figure 1.14 shows charge characterization of an IMAC-purified membrane protein using Mono Q™.

Figure 1.14.Charge homogeneity characterization of histidine-tagged cytochrome bo3 ubiquinol oxidase using anion exchange chromatography with Mono Q. Two fractions obtained from IMAC purification were analyzed. Electrophoretic analysis was done with PhastSystemTM using PhastGelTM 8-25% and silver staining. M = Low Molecular Weight Calibration Kit; E = detergent extract of E. coli membranes; FT = flowthrough material; F1 = fraction 1 from HiTrap Chelating HP 1 mL; F2 = fraction 2 from HiTrap Chelating HP 1 mL. See application note 18-1128-92 Purification and chromatographic characterization of an integral membrane protein.”
*Determined for soluble, globular proteins.
The solubility of membrane proteins can be very sensitive to ionic strength. When this is the case, fractions eluted in a salt gradient from an ion exchange chromatography column should be immediately diluted or run on a PD-10 Desalting, HiTrap Desalting, or a HiPrep 26/10 Desalting column to reduce the salt concentration (e.g., to < 50 mM).
Gel filtration is excellent for the final purification step as it both removes aggregates and simultaneously achieves buffer exchange, if required.
Tag cleavage
The affinity tag can be removed after purification if a protease cleavage site has been inserted between the tag and the target protein. If using an ÄKTAxpress system, on-column cleavage protocols can be performed automatically. Protease activity can be affected by the presence of detergents. Comprehensive data on the activity of different proteases in detergents is largely lacking, but both PreScission™ Protease and thrombin have been used successfully with detergents. It is recommended to check for the extent of cleavage under different conditions by SDS-PAGE.
PreScission protease exhibits excellent cleavage properties at 4 °C and is a useful alternative for cleavage of sensitive proteins.
TEV protease is fully active in 9 mM decyl maltoside, but is partly inactive in several other detergents.
Materials
如要继续阅读,请登录或创建帐户。
暂无帐户?