GenElute™ RNA/DNA/Protein Purification Kit
Product No. E5163
Product Description
Our GenElute™ RNA/DNA/Protein Plus Purification Kit provides a rapid method for the isolation and purification of total RNA, genomic DNA and proteins sequentially from a single sample of cultured animal cells, small tissue samples, blood, bacteria, yeast, fungi or plants. The total RNA, genomic DNA and proteins are all column purified in less than 30 minutes.
This kit is ideal for researchers who are interested in studying the genome, proteome and transcriptome of a single sample, such as for studies of microRNA profiling, gene expression including gene silencing experiments or mRNA knockdowns, studies involving biomarker discovery, and for characterization of cultured cell lines. This kit is especially useful for researchers who are isolating macromolecules from precious, difficult to obtain or small samples such as biopsy materials or single foci from cell cultures, as it eliminates the need to fractionate the sample. Furthermore, analysis will be more reliable since the RNA, DNA and proteins are derived from the same sample, thereby eliminating inconsistent results. The purified macromolecules are of the highest purity and can be used in a number of different downstream applications.
The kit purifies all sizes of RNA, from large mRNA and ribosomal RNA down to microRNA (miRNA) and small interfering RNA (siRNA). The purified RNA is of the highest integrity and can be used in a number of downstream applications including real time PCR, reverse transcription PCR, Northern blotting, RNase protection and primer extension, and expression array assays. The genomic DNA is of the highest quality, and can be used in PCR reactions, sequencing, Southern blotting and SNP analysis.
Product Overview
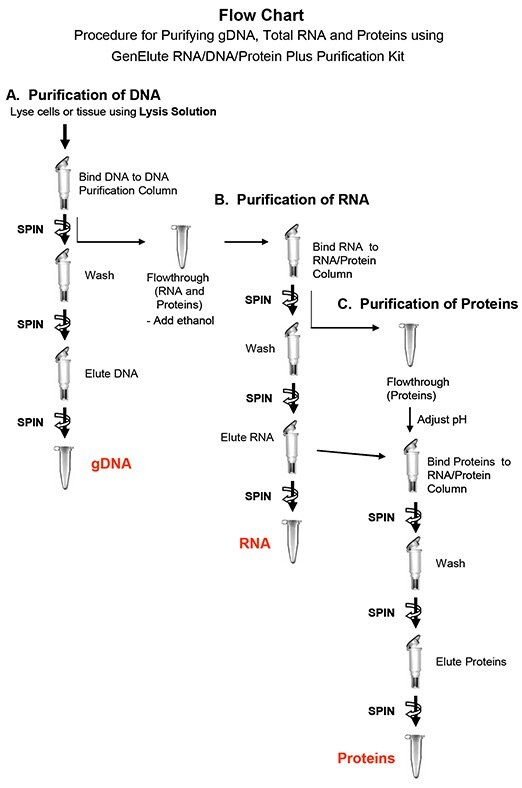
DNA and RNA purification is based on spin column chromatography. The process involves first lysing the cells or tissue of interest with the provided Lysis Solution. The DNA is then captured and purified on a DNA Purification Column. Ethanol is then added to the flowthrough of the DNA purification step, and the solution is loaded onto a RNA/Protein Purification Column. The resin in the column binds nucleic acids in a manner that depends on ionic concentrations, thus only the RNA including microRNAs will bind to the column while the proteins are removed in the flowthrough. Next, the bound RNA is washed with the provided RNA Wash Solution to remove impurities, and the purified RNA is eluted with the RNA Elution Solution.
The proteins that are present from the RNA binding flowthrough can now be loaded directly onto an SDSPAGE gel for visual analysis. Alternatively, the protein samples can be further purified using the same RNA/Protein Purification Column that was used for purifying the RNA. After the RNA has been eluted from the column, the flowthrough is then pH adjusted and loaded back onto the column in order to bind the proteins that are present. The bound proteins are washed with the provided wash buffer, and are then eluted such that they can be used in downstream applications.
Precautions and Disclaimer
This product is for R&D use only, not for drug, household, or other uses. Please consult the Safety Data Sheet for information regarding hazards and safe handling practices.
This kit contains solutions that may contain irritants. Wear gloves and safety glasses when handling any reagent provided in this kit.
Storage/Stability
This kit is shipped in two parts: 1) Cat. No. E5164-1KT, includes reagents shipped at room temperature, and 2) Cat. No. 23506NB, Protein Loading Dye, is shipped on dry ice. The Protein Loading Dye should be stored at -20 °C upon arrival. All other solutions should be kept tightly sealed and stored at room temperature.
Reagents and equipment required, not provided
For All Protocols
- Benchtop microcentrifuge
- 2-mercaptoethanol (M3148)
- 96-100% ethanol (E7148, E7023, or 459836)
- Isopropanol (I9516 or 59304)
- Molecular biology grade water (W4502)
For Animal Cell Protocol
- PBS (RNase-free), (P5493)
For Animal Tissue Protocol
- Liquid nitrogen
- Mortar and pestle
For Bacterial Protocol
- Lysozyme-containing TE Buffer:
For Yeast Protocol
- Resuspension Buffer with Lyticase:
- 50 mM Tris pH 7.4
- 10 mM EDTA
- 1 M Sorbitol
- 1 unit/µL Lyticase (L2524)
For Fungi Protocol
- Liquid nitrogen
- Mortar and pestle
For Plant Protocol
- Liquid nitrogen
- Mortar and pestle
Working with RNA
RNases are very stable and robust enzymes that degrade RNA. Autoclaving solutions and glassware is not always sufficient to actively remove these enzymes. The first step when preparing to work with RNA is to create an RNase-free environment. The following precautions are recommended as your best defense against these enzymes.
- The RNA area should be located away from microbiological work stations
- Clean, disposable gloves should be worn at all times when handling reagents, samples, pipettes, disposable tubes, etc. It is recommended that gloves are changed frequently to avoid contamination
- There should be designated solutions, tips, tubes, lab coats, pipettes, etc. for RNA only
- All RNA solutions should be prepared using at least 0.05% DEPC-treated autoclaved water or molecular biology grade nuclease-free water
- Clean all surfaces with commercially available RNase decontamination solutions, such as RNase AWAY® (83931)
- When working with purified RNA samples, ensure that they remain on ice during downstream applications
Preparation Instructions
Before beginning the procedure, complete the following:
- Ensure that all solutions are at room temperature prior to use.
- RNA Wash Solution: Add 50 mL of 96-100% ethanol to the concentrated RNA Wash Solution. This will give a final volume of 72 mL. Mark the label of the bottle to indicate that the ethanol is added.
- gDNA Wash Solution I: Add 20 mL of 96 - 100% ethanol to the concentrated gDNA Wash Solution I. This will give a final volume of 30 mL. Mark the label of the bottle to indicate that the ethanol is added.
- gDNA Wash Solution II: Add 15 mL of 96-100% ethanol to the concentrated gDNA Wash Solution II. This will give a final volume of 30 mL. Mark the label of the bottle to indicate that the ethanol is added.
- Optional: The use of ß-mercaptoethanol in lysis is highly recommended for most tissues, particularly those known to have high RNase content (ex: pancreas). It is also recommended for researchers who wish to isolate RNA for sensitive downstream applications. Add 10 µL of 2- mercaptoethanol to each 1 mL of Lysis Solution. Alternatively, the lysis solution can be used as provided.
Procedure
I. Sample Preparation
Cell Preparation
For optimal results, it is recommended that 1 x 106 cells be used for the input. As a general guideline, a confluent 3.5 cm plate of HeLa cells will contain 106 cells. Inputs of up to 5 x 106 cells may be used; however slight cross-contamination of genomic DNA in the RNA fraction may be observed in input ranges greater than 106 cells.
- Harvest cells.
- Attached cell cultures: Attached cells can be lysed directly in culture vessel. Wash the cells in PBS.
- Suspension and lifted cells: Pellet up to to 5 x 106 cells for 10 minutes at ~ 200 x g. Carefully, decant the supernatant to ensure that the pellet is not dislodged. Wash the cell pellet with an appropriate amount of PBS. Centrifuge at 200 x g (~2,000 RPM) for another 5 minutes. Carefully decant the supernatant. Several microliters (µL) of PBS may be left behind with the pellet in order to ensure that the pellet is not dislodged.
Note: Cell pellets can be stored at -70 °C for later use or used directly in the procedure. Determine the number of cells present before freezing. Frozen pellets should be stored for no longer than 2 weeks to ensure that the integrity of the RNA is not compromised. Frozen cell pellets should not be thawed prior to beginning the protocol. Add the Lysis Solution directly to the frozen cell pellet (Lysis Step 2).
- Lyse cells.
- Attached cell cultures: Add 300 µL of Lysis Solution directly to culture plate. Lyse cells by gently tapping culture dish and swirling buffer around plate surface for five minutes. Transfer lysate to a microcentrifuge tube. Proceed to Section II.
- Suspension and lifted cells: Add 300 µL of Lysis Solution to the pellet. Lyse cells by vortexing for 15 seconds. Ensure that the entire pellet is completely dissolved before proceeding to the next step. Proceed to Section II.
Note: For input amounts greater than 106 cells, it is recommended that the lysate is passed through a 25 gauge needle attached to a syringe 5-10 times at this point, in order to reduce the viscosity of the lysate prior to loading onto the column.
Animal Tissues Preparation
RNA in animal tissues is not protected after harvesting until it is disrupted and homogenized. Thus it is important that the procedure is carried out as quickly as possible, particularly the Cell Lysate Preparation step.
Fresh or frozen tissues may be used for the procedure. Tissues should be flash-frozen in liquid nitrogen and transferred immediately to a -70 °C freezer for long-term storage. Tissues may be stored at -70 °C for several months. Do not allow frozen tissues to thaw prior to grinding with the mortar and pestle in order to ensure that the integrity of the RNA is not compromised.
The maximum recommended input of tissue varies depending on the type of tissue being used. Please refer to Table 1 below as a guideline for maximum tissue input amounts. If your tissue of interest is not included in the table below, we recommend starting with an input of no more than 10 mg.
- Prepare tissue.
- Excise the tissue sample from the animal.
- Determine the amount of tissue by weighing. Please refer to Table 1 for the recommended maximum input amounts of different tissues. For tissues not included in the table, we recommend starting with an input of no more than 10 mg.
- Transfer the tissue into a mortar that contains an appropriate amount of liquid nitrogen to cover the sample. Grind the tissue thoroughly using a pestle.
- Allow the liquid nitrogen to evaporate, without allowing the tissue to thaw.
- Lyse tissue.
- Add 300 µL of Lysis Solution to the tissue sample and continue to grind until the sample has been homogenized. Homogenize by passing the lysate 5-10 times through a 25 gauge needle attached to a syringe.
- Using a pipette, transfer the lysate into an RNase-free microcentrifuge tube (not provided).
- Spin lysate for 2 minutes to pellet any cell debris. Transfer the supernatant to another RNase-free microcentrifuge tube (not provided). Note the volume of the supernatant/lysate. Proceed to Section II.
Lysate Preparation from Blood
Blood of all human and animal subjects is considered potentially infectious. All necessary precautions recommended by the appropriate authorities in the country of use should be taken when working with whole blood.
It is recommended that no more than 100 µL of blood be used in order to prevent clogging of the column. We recommend the use of this kit to isolate RNA from noncoagulating fresh blood using EDTA as the anticoagulant.
- Transfer up to 100 µL of non-coagulating blood to an RNase-free microcentrifuge tube (not provided).
- Add 300 µL of Lysis Solution to the blood. Lyse cells by vortexing for 15 seconds. Ensure that mixture becomes transparent before proceeding to the next step. Proceed to Section II.
Lysate Preparation from Bacteria
Prepare the appropriate lysozyme-containing TE Buffer as indicated in Table 2. This solution should be prepared with sterile, RNAse-free TE Buffer, and kept on ice until needed. These reagents are to be provided by the user.
It is recommended that no more than 109 bacterial cells be used in this procedure. Bacterial growth can be measured using a spectrophotometer. As a general rule, an E. coli culture containing 1 x 109 cells/mL has an OD600 of 1.0. For RNA isolation, bacteria should be harvested in log-phase growth. Bacterial pellets can be stored at -70 °C for later use, or used directly in this procedure.
- Bacteria harvest.
- Pellet bacteria by centrifuging at 14,000 x g (~14,000 RPM) for 1 minute.
- Decant supernatant, and carefully remove any remaining media by aspiration. The bacterial pellet can be frozen at -70 °C for later use, or used immediately.
- Bacterial cell lysis.
- Resuspend the bacteria thoroughly in 100 µL of the appropriate lysozyme-containing TE Buffer (Table 2) by vortexing. Incubate at room temperature for the time indicated in Table 2. If using frozen bacterial pellets, do not thaw prior to beginning the protocol. Add the Lysozyme-containing TE Buffer directly to the frozen bacterial pellet.
- Add 300 µL of Lysis Solution and vortex vigorously for at least 10 seconds. Proceed to Section II.
Lysate Preparation from Yeast
Prepare the appropriate amount of Lyticase-containing Resuspension Buffer, considering that 500 µL of buffer is required for each preparation. The Resuspension Buffer should have the following composition: 50 mM Tris, pH 7.5, 10 mM EDTA, 1M Sorbitol, 0.1% ß-mercaptoethanol and 1 unit/µL Lyticase. This solution should be prepared with sterile, RNAse-free reagents, and kept on ice until needed. These reagents are to be provided by the user.
It is recommended that no more than 107 yeast cells or 1 mL of culture be used for this procedure. For RNA isolation, yeast should be harvested in log-phase growth.
Yeast can be stored at -70 °C for later use, or used directly in this procedure. Frozen yeast pellets should not be thawed prior to beginning the protocol. Add the Lyticase-containing Resuspension Buffer directly to the frozen yeast pellet (Lysis Step 2).
Cell Lysate Preparation from Yeast
- Yeast cell harvest.
- Pellet yeast by centrifuging at 14,000 x g (~14,000 RPM) for 1 minute.
- Decant supernatant, and carefully remove any remaining media by aspiration.
- Resuspend the yeast thoroughly in 500 µL of Lyticase-containing Resuspension Buffer by vortexing. Incubate at 37 °C for 10 minutes.
- Pellet the spheroplasts at 200 x g (~2,000 RPM) for 3 minutes. Decant supernatant.
- Lysis of spheroplasts. Add 300 µL of Lysis Solution and vortex vigorously for at least 10 seconds. Proceed to Section II.
Lysate Preparation from Fungi
Fresh or frozen fungi may be used for this procedure. Fungal tissue should be flash-frozen in liquid nitrogen and transferred immediately to a -70 °C freezer for long-term storage. Fungi may be stored at -70 °C for several months. Do not allow frozen tissues to thaw prior to grinding with the mortar and pestle in order to ensure that the integrity of the RNA is not compromised. It is recommended that no more than 50 mg of fungi be used for this procedure in order to prevent clogging of the column. It is important to work quickly during this procedure.
Cell Lysate Preparation from Fungi
- Fungi harvest and cell preparation.
- Determine the amount of fungi by weighing. It is recommended that no more than 50 mg of fungi be used for the protocol.
- Transfer the fungus into a mortar that contains an appropriate amount of liquid nitrogen to cover the sample. Grind the fungus thoroughly using a pestle.
Note: At this stage the ground fungus may be stored at -70 °C, such that the RNA purification can be performed at a later time. - Allow the liquid nitrogen to evaporate, without allowing the tissue to thaw.
- Lyse cells.
- Add 300 µL of Lysis Solution to the tissue sample and continue to grind until the sample has been homogenized.
- Using a pipette, transfer the lysate into an RNase-free microcentrifuge tube (not provided).
Spin lysate for 2 minutes to pellet any cell debris. Transfer the supernatant to another RNase-free microcentrifuge tube. Note the volume of the supernatant/lysate. Proceed to Section II.
Lysate Preparation from Plant
The maximum recommended input of plant tissue is 50 mg or 5 x 106 plant cells. Both fresh and frozen plant samples can be used for this protocol. Samples should be flash-frozen in liquid nitrogen and transferred immediately to a -70 °C freezer for longterm storage. Do not allow frozen tissues to thaw prior to grinding with the mortar and pestle in order to ensure that the integrity of the RNA is not compromised.
Cell Lysate Preparation from Plant
- Plant cell preparation
- Transfer ≤50 mg of plant tissue or 5 x 106 plant cells into a mortar that contains an appropriate amount of liquid nitrogen to cover the sample. Grind the sample into a fine powder using a pestle in liquid nitrogen.
Note: If stored frozen samples are used, do not allow the samples to thaw before transferring to the liquid nitrogen. - Allow the liquid nitrogen to evaporate, without allowing the tissue to thaw.
- Transfer ≤50 mg of plant tissue or 5 x 106 plant cells into a mortar that contains an appropriate amount of liquid nitrogen to cover the sample. Grind the sample into a fine powder using a pestle in liquid nitrogen.
- Lyse plant cells
- Add 600 µL of Lysis Solution to the tissue sample and continue to grind until the sample has been homogenized.
- Using a pipette, transfer the lysate into an RNase-free microcentrifuge tube (not provided).
Spin lysate for 2 minutes to pellet any cell debris. Transfer the supernatant to another RNase-free microcentrifuge tube. Note the volume of the supernatant/lysate. Proceed to Section II.
II: Genomic DNA Purification
Note: The following steps of the procedure for the purification of genomic DNA are the same for all the different types of lysate.
- Binding DNA to gDNA Purification Column
- Retrieve a gDNA Purification Column preassembled with a collection tube.
- Apply up to 600 µL of the lysate prepared from Section 1 onto the column and centrifuge at 14,000 x g for 1 minute. Ensure the entire lysate volume has passed through into the collection tube by inspecting the column. If the entire lysate volume has not passed, spin for an additional minute.
- Retain the flowthrough for RNA Purification (Section III). The flowthough contains the RNA and proteins and should be stored on ice or at -20 °C until the RNA Purification protocol is carried out.
- Reassemble the spin column with the collection tube.
- Genomic DNA Wash
- Apply 500 µL of gDNA Wash Solution I to the column and centrifuge at 14,000 x g for 1 minute. Discard the flowthrough.
- Apply 500 µL of gDNA Wash Solution II to the column and centrifuge at 14,000 x g for 1 minute. Discard the flowthrough.
- Spin the column at 14,000 x g for 2 minutes in order to thoroughly dry the resin. Discard the collection tube.
- Genomic DNA Elution
- Place the column into a fresh 1.7 mL Elution tube provided with the kit.
- Add 100 µL of gDNA Elution Buffer to the column.
- Centrifuge for 2 minutes at 200 x g, followed by 1 minute at 14,000 x g. Note the volume eluted from the column. If the entire volume has not been eluted, spin the column at 14,000 x g for 1 additional minute.
- Storage of DNA
The purified DNA sample may be stored at 4 °C for a few days. It is recommended that samples be placed at ≤ –20 °C for long term storage.
III: Total RNA Purification
Note: For sensitive applications that require the complete removal of genomic DNA, an optional on-column DNase I treatment could be performed after completion of Step 1.6.
- Binding RNA to Column
- To every 100 µL of flowthrough from Step 2.3, add 60 µL of 96-100 % ethanol. Mix by vortexing.
Note: For example, for 300 µL of flowthrough, add 180 µL of 96-100 % ethanol - Assemble an RNA/Protein Purification Column with one of the provided collection tubes.
- Apply up to 600 µL of the lysate with the ethanol onto the column and centrifuge for at 14,000 x g for 1 minute. Ensure that the entire lysate volume has passed through into the collection tube by inspecting the column. If the entire lysate volume has not passed, spin for an additional minute.
- Retain the flowthrough for Protein Purification (Section IV). The flowthrough contains the proteins and should be stored on ice or at -20 °C until the Protein Purification protocol is carried out.
- Depending on your lysate volume, repeat steps 1.3 and 1d if necessary. The flowthroughs should be combined and retained in the same microcentrifuge tube.
- Reassemble the spin column with the collection tube.
- To every 100 µL of flowthrough from Step 2.3, add 60 µL of 96-100 % ethanol. Mix by vortexing.
- RNA Wash
- Apply 400 µL of RNA Wash Solution to the column and centrifuge at 14,000 x g for 1 minute.
Note: Ensure the entire wash solution has passed through into the collection tube by inspecting the column. If the entire wash volume has not passed, spin for an additional minute. - Discard the flowthrough and reassemble the column with the collection tube.
- Wash the column a second time by adding another 400 µL of RNA Wash Solution and centrifuge at 14,000 x g for 1 minute.
- Discard the flowthrough and reassemble the spin column with its collection tube.
- Wash the column a third time by adding another 400 µL of RNA Wash Solution and centrifuge at 14,000 x g for 1 minute.
- Discard the flowthrough and reassemble the spin column with its collection tube.
- Spin the column at 14,000 x g for 2 minutes in order to thoroughly dry the resin. Discard the collection tube.
- Apply 400 µL of RNA Wash Solution to the column and centrifuge at 14,000 x g for 1 minute.
- RNA Elution
- Place the column into a fresh 1.7 mL Elution tube provided with the kit.
- Add 50 µL of RNA Elution Solution to the column.
- Centrifuge for 2 minutes at 200 x g, followed by 1 minute at 14,000 x g. Note the volume eluted from the column. If the entire volume has not been eluted, spin the column at 14,000 x g for 1 additional minute.
Note: For maximum RNA recovery, particularly for samples that are know to contain large amounts of RNA, it is recommended that a second elution be performed into a separate microcentrifuge tube (Repeat Steps 3.2 and 3.3). - Retain the column for Protein Purification. Proceed to Section IV for Protein Purification.
- Storage of RNA
The purified RNA sample may be stored at –20 °C for a few days. It is recommended that samples be placed at –70 °C for long term storage.
IV. Procedure to Isolate Total Proteins from All Cell Types
Notes Prior to Use
- At this point, the proteins that are present in the flowthrough from the RNA Binding Step (Section III, Step 1.4) can be processed by one of the following three options:
- Direct running on an SDS-PAGE gel with the provided loading dye for visual analysis
- Column purification (recommended)
- Acetone precipitation
- For direct running on a gel, the provided Protein Loading Dye should be used instead of regular SDS-PAGE Loading Buffer in order to prevent any precipitates from forming. Add 1 volume of the Protein Loading Dye to the sample and boil for 2 minutes before loading.
- Column purification of the proteins is recommended.
- For acetone precipitation, please refer to the supplementary protocol provided in the Appendix A below.
- pH Adjustment of Lysate
- Transfer the flowthrough from the RNA Binding Step (Section III, Step 1.4) to a separate microcentrifuge tube.
- For every 100 µL of flowthrough, dilute with 100 µL of molecular biology grade water.
Note: For example, to purify the entire flowthrough of 480 µL, dilute with 480 µL molecular biology grade water. - For every 100 µL of flowthrough, add 8 µL (or 40 µL for an entire flowthrough of 480 µL) of Protein pH Binding Buffer. Mix contents well.
Note: Depends on the type and amount of input, slight precipitation may occur which will not affect the purification procedure.
- Protein Binding
- Apply up to 600 µL of the pH-adjusted protein sample onto the column, and centrifuge for 2 minutes at 5,200 x g (~8,000 RPM). Inspect the column to ensure that the entire sample has passed through into the collection tube. If necessary, spin for an additional 3 minutes.
- Discard the flowthrough. Reassemble the spin column with its collection tube.
Note: You can save the flowthrough in a fresh tube for assessing your protein’s binding efficiency. - Depending on your sample volume, repeat steps 2.1 and 2.2 until the entire protein sample has been loaded onto the column.
- Column Wash
- Apply 500 µL of Protein Wash Buffer to the column and centrifuge for 2 minutes at 5,200 x g.
- Discard the flowthrough and reassemble the spin column with its collection tube.
- Inspect the column to ensure that the liquid has passed through into the collection tube. There should be no liquid in the column. If necessary, spin for an additional minute to dry.
- Protein Elution and pH Adjustment
The supplied Protein Elution Buffer consists of 10 mM sodium phosphate pH 12.5.- Add 9.3 µL of Neutralizer to a fresh 1.7 mL Elution tube.
- Transfer the spin column from the Column Wash procedure into the Elution tube.
- Apply 100 µL of the Protein Elution Buffer to the column and centrifuge for 2 minutes at 5,200 x g to elute bound proteins.
Appendix A: Acetone Precipitation Procedure for Proteins
- Add 4 volumes of ice-cold acetone to the flowthrough from the RNA Binding Step (Section 3, Step 6.4).
- Incubate for 15 minutes on ice or at -20 °C.
- Centrifuge for 10 minutes at 14,000 x g (~12,000 RPM). Discard the supernatant and allow the pellet to air-dry.
Note: At this point the pellet can be washed with 100 mL of ice cold ethanol and again air-dried. - Resuspend the pellet in the buffer of your choice that is suited to your downstream application.
Troubleshooting Guide
GenElute is a registered trademark of Merck KGaA, Darmstadt, Germany and/or its affiliates.
如要继续阅读,请登录或创建帐户。
暂无帐户?