High-Throughput PROTAC Synthesis: Kits for Targeted Protein Degradation
Introduction
PROTACs (Proteolysis Targeting Chimeras) represent a groundbreaking approach in targeted protein degradation, leveraging the ubiquitin-proteasome system to selectively eliminate unwanted proteins within cells. By designing bifunctional molecules that bind both the target protein and the E3 ligase, PROTACs facilitate the recruitment of the proteasome to the target protein, leading to its ubiquitination and subsequent degradation. This targeted approach not only enhances the specificity of protein degradation but also minimizes off-target effects, making PROTACs a promising avenue for drug development.
This innovative technology holds immense potential for therapeutic applications, particularly in oncology and other diseases where protein dysregulation is a key factor.1 Additionally, PROTACs have applications in other diseases characterized by protein misfolding or aggregation, such as neurodegenerative disorders. As research in this area continues to evolve, PROTACs are expected to pave the way for novel therapeutic strategies that can address previously undruggable targets, ultimately transforming the landscape of drug discovery and development.
The QuicTPD™ Acid and QuicTPD™ Amine Screening Sets are collections of 36 partial PROTACs/E3-ligase–linker conjugates, designed with diverse linkers of varying architectures and lengths, with the acid set featuring a carboxylic acid terminal and the amine set an amine terminal. This set is ideal for researchers looking to develop proof-of-concept PROTAC degraders for their targets of interest. It specifically targets Cereblon (CRBN) and von Hippel-Lindau (VHL) E3 ligases, both of which play crucial roles in the ubiquitin-proteasome system and are pivotal in PROTAC discovery for targeted protein degradation (TPD).

Figure 1.Visual overview of the assembly of QuicTPD™ Acid/Amine Screening Set. 1a is a sealing mat, 1b is a TPU vial downloader, 1c is a glass shell vial with partial PROTACs in µmole quantities, and 1d is a vial holder.
Each screening set contains the following components:
- The QuicTPD™ Acid/Amine Screening Set includes 36 partial PROTACs (see Table 1), plated in duplicate across 72 glass shell vials. Each glass shell vial contains either 1.5 micromoles or 5 micromoles or partial PROTAC based on the SKU size of 3 micromoles or 10 micromoles, respectively.
- Silicon Mat (PM32121) for covering glass shell vials.
- The TPU (thermoplastic urethane) downloader (PM72334) allows for parallel handling of all glass shell vials.
- Vial holder for holding vials.
- 24 empty vials for setting up controls and/or reaction optimization.
Table 1: Partial PROTACs in the QuicTPD™ Acid/Amine Screening Set
Partial PROTACs in the QuicTPD™ Acid Screening Set
Partial PROTACs in the QuicTPD™ Amine Screening Set
Features and Benefits
- The QuicTPD™ Acid/Amine Screening Set enables seamless peptide coupling reactions with warheads that possess solvent-exposed, reactive handles, such as acids or amines, allowing efficient assembly of PROTACs targeting proteins of interest.
- Each partial PROTAC (E3 ligase + linker) is conveniently plated in micromole scale within glass shell vials, facilitating a ready-to-react workflow that accommodates various reaction conditions, including different coupling reagents, bases, and temperatures.
- With the ability to react with at least two distinct warheads, researchers can create 72 or more unique small molecule degraders for further screening in a direct-to-biology (D2B) approach2-6. This miniaturized synthesis approach not only accelerates PROTAC discovery but also promotes sustainability by reducing the use of solvents, reagents, water, and energy. (Figures 2a, 2b, and 2c).
- Partial PROTACs are based on Lenalidomide, Pomalidomide, Piperidinedione, and VH032 with a rich diversity in linkers for hydrophilicity, length, and rigidity to maximize your SAR discovery campaign.
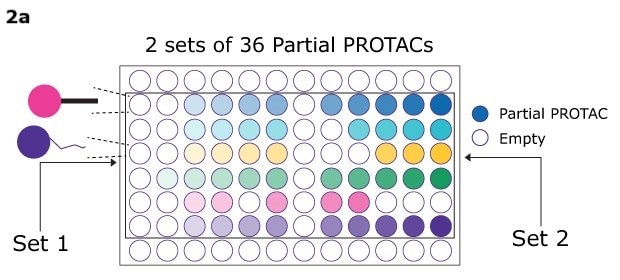
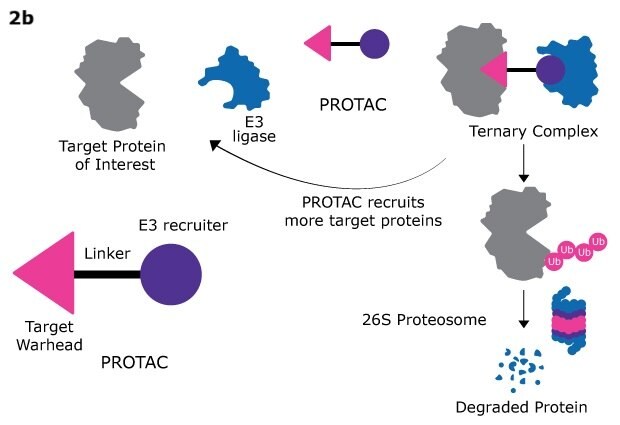
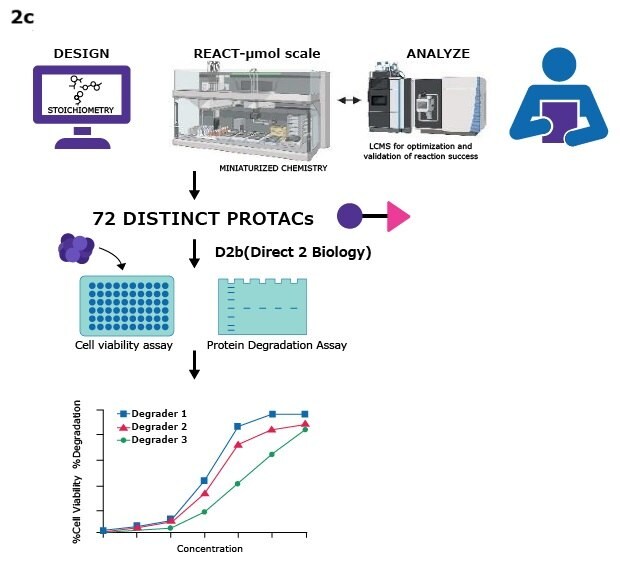
Figure 2a. 2 sets of 36 partial PROTACs, 2b. targeted protein degradation via PROTACs, 2c. general workflow for assembling PROTACs using the QuicTPD™ acid/amine screening set
PROTAC Library Generation with QuicTPD™ Acid/Amine Screening Set
The following general protocol is recommended for constructing PROTAC libraries through peptide coupling reactions. This involves the reaction between warheads targeting specific proteins with pendant acid/amine reactive terminals, which can effectively couple with the amine or acid partial PROTACs available in the QuicTPD™ Screening Sets.
Required Reagents and Setup Essentials
Warheads: It is advisable to use at least 2 warheads to generate 72 PROTAC degraders through combinatorial synthesis with the 36 pre-plated partial PROTACs at a given SKU size of 3 and 10 micromoles. D2B reactions can very well be carried out at the nanomole scale; hence, the contents of the plates can be dissolved and aliquoted into multiple plates to react with the desired number of warheads with complimentary reactive termini. Ensure that the warheads are generally inert in downstream screening assays, especially if a direct-to-biology approach is employed.
Coupling agents: The following commonly used reagents are essential for typical peptide coupling reactions. These are not included with the screening set and should be purchased separately.
- EDC 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (39391)
- NMM (N-Methylmorpholine) as a base (M56557)
- Oxyma Pure as an additive (8.51086)
Solvents: Dimethyl sulfoxide (DMSO) (1.02931) is typically used in peptide coupling reactions. This solvent is not provided with the screening set and should be acquired separately.
Temperature, time, and mixing parameters: Conduct the reaction at room temperature (25°C) for 12- 24 hours on a horizontal shaker using standard settings. For more challenging peptide coupling reactions, the reaction vials can be transferred to aluminum heating block reactors.
Scale of reaction: The reaction scale typically varies from nanomole to micromole.
Final concentration of PROTAC: The desired final concentration typically ranges from 1 to 10 mM.
Final volume of reaction (50-500 microliters): It is recommended to keep low reaction volumes to ensure enough headspace in reaction vials to prevent cross-contamination between reactions.
Concentration limits of coupling agents: It is recommended to keep EDC, Oxyma Pure, and NMM concentrations in reaction at or below 175 mM, 500 mM, and 240 mM, respectively, for direct biology screening.
Refer to the papers listed in the References section for further reading on reaction conditions.
Detailed Protocol
- Stoichiometric calculations: For assistance with stoichiometric calculations, please refer to the Cal sheet.
- Weighing reagents: Accurately weigh the required amounts of warheads, acid activator, additive, and base into glass vials.
- Preparation of stock solutions: Prepare stock solutions of the acid activator, additive, base, and warheads in your solvent of choice, preferably anhydrous DMSO.
- Prepping up the reaction plate: Add the reaction solvent in the necessary quantities to the glass shell vials containing the partial PROTACs. Mix the contents by aspiration and shaking for 30 minutes to ensure proper dissolution.
- Aliquoting for smaller scales: If the user is conducting reactions below SKU size, aliquot the required volume into daughter plates for small-scale reactions.
- Addition of warheads and coupling reagents: It is recommended to pre-activate the acid warheads with the coupling reagent. However, you can also sequentially add the required quantities of warheads, followed by the acid activator, additives, and base to achieve the desired final concentration and reaction volume. Generally, the ratios should be warhead:partial PROTAC:coupling reagent:additive:base = 1:1.2-1.5:1.5:2:8.0.
- Setting up controls: Use empty glass shell vials to set up negative controls to assess whether the reaction components themselves affect downstream assays. Examples of negative controls that can be included in technical replicates are warheads only, warheads and coupling agents, coupling agents only, and solvent only. These controls are recommended if a direct-to-biology approach is employed.
- Reaction time, temperature, and mixing: Peptide coupling reactions are typically conducted for 12 to 24 hours at room temperature. For optimal results, place the reaction plates securely on a horizontal shaker overnight at low to medium speed. To minimize exposure of the formed PROTACs to moisture and light, ensure that the silicone mat is properly secured on each glass shell vial to prevent cross-contamination.
- Post-reaction procedures: After the reaction is complete, transfer an aliquot of the contents from the plate to a separate 96-well plate for analysis of warhead conversion. Warhead consumption can be assessed using HPLC/MS to evaluate reaction efficiency. The remaining contents of the plate, including the PROTACs formed in situ, can be stored at -20°C for further downstream analysis. Ensure that the silicone mat is secured on each glass shell vial to limit exposure to moisture and light. Align the A1 corner of the vial holder with the A1 mark on the silicon mat. Proper alignment is required to ensure the probe maps correctly to each vial and to avoid cross-contamination.
Other Products of Interest
QuicTPD™ VHL and CRBN ligand sets (QVHLSET-1KT and QCRBNSET-1KT) are specifically designed for researchers who already have warhead ligated to linkers targeting their proteins of interest. With these sets, researchers can utilize the E3 recruiters provided in the QVHLSET and QCRBNSET to construct a diverse library of PROTACs tailored to their research needs. Once a researcher identifies a small molecule degrader of interest, they can further enhance their studies using the QuicTPD™ Negative Control Set (QNEGCONSET-1KT). This set includes CRBN and VHL recruiters with slightly varied structures, allowing for the construction of PROTACs that are not recognized by E3 ligases. These negative controls are essential for confirming the mechanism of action of the identified degraders. Additionally, to validate that degradation occurs via the ubiquitin-proteasome pathway, researchers can utilize MG132 (M8699), a potent proteasome inhibitor, and MLN4924 (5.05477), a neddylation inhibitor. Together, these tools will enable researchers to comprehensively explore and confirm their findings in targeted protein degradation.
The QuicTPD™ Acid/Amine Optimization set is designed to facilitate high-throughput synthesis optimization for researchers working with PROTACs. This set includes 4 representative partial PROTACs, each provided at a 5 micromole quantity. Two of these PROTACs are tailored to target the CRBN E3 ligase, while the other two are directed towards VHL, incorporating both hydrophobic and hydrophilic linkers. By utilizing this optimization set, researchers can explore a variety of reaction conditions, up to 24 or more, before proceeding to the complete screening set. This exploration includes testing different coupling reagents, solvents, and bases, allowing for a comprehensive assessment of the synthesis parameters that influence the efficiency and yield of the desired products.
In this article, we use the term ‘PROTAC’ as the common abbreviation for the therapeutic modalities encompassed by PROteolysis TArgeting Chimera. PROTAC® is a registered trademark of Arvinas Operations, Inc.
Related Products
Related Webinars

Join our webinar to discover the innovative QuicTPD™ 'direct-to-biology' platform, which accelerates PROTAC drug discovery from chemical design to biological evaluation
References
如要继续阅读,请登录或创建帐户。
暂无帐户?