Lysing Enzymes
- Cell lysis enzymes – how to tear down the wall
- Understanding the bacterial cell wall – the gram positives and negatives
- Which enzymes will get the job done – lysing bacteria
- The microbiome - sequencing and phylogeny
- DNA-free lytic enzymes for studying the microbiome
- The yeast cell wall – shape and rigidity
- Enzymes that can break down the yeast cell wall
- Toxins for eukaryotic cells
- Making eukaryotic cells permeable – with a toxin
- Enzymes for plant cell lysis
Cell Lysis Enzymes – How to Tear Down the Wall
Enzymes provide a non-mechanical method for cell lysis and protoplast preparation. It may seem like a simple process to throw in your enzyme, stick your tube in the water-bath and walk away, but what is actually going on in that process? What is being disrupted and how can you choose the best enzyme for the job?
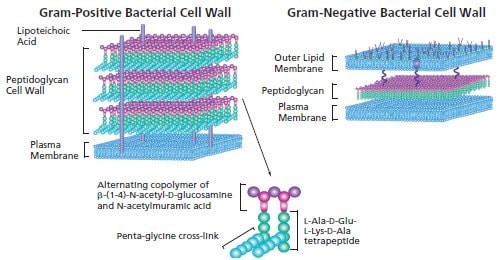
Figure 1.Gram Positive Bacterial Cell Wall and Gram Negative Bacterial Cell Wall
Gram-positive bacteria is composed of multiple layers of peptidoglycan. Peptioglycan is a Polymer of β-(1-4)-N-Acetyl-D-glucosamine units. Alternating residues are modified to form N-acetylmuramic acid with the addition of lactate to form branching links to a tetrapeptide. The tetrapeptides of adjacent polymers are linked by penta-glycine bridges. The cross-linked peptidoglycan polymers form a mesh-like network over a phospholipid bylayer plasma membrane.
Gram-negative cell wall in bacteria is composed of an outer lipid bylayer, which, in addition to phospholipids, is also covered with lipopolysaccharide moieties. Lipoproteins link the outer lipid membrane to the thin peptidoglycan layer in the periplasmic space. The inner plasma membrane is a phospholipid bilayer.
Which Enzymes Will Get the Job Done - Lysing Bacteria
Labiase
Product Number L1414 (powder) From Streptomyces fulvissimus
- Labiase from Streptomyces fulvissimus is an enzyme preparation useful for the lysis of many Gram-positive bacteria such as Lactobacillus, Aerococcus and Streptococcus.
- Labiase contains β-N-acetyl-D-glucosaminidase and lysozyme activity.
- pH Optimum for activity: pH ~ 4
- pH Optimum for stability: pH ~ 4-8
Lysostaphin
Product Number L7386 (lyophilized powder, min. 500 units/mg protein)
Product Number L4402 (lyophilized powder, min. 2,000 units/mg protein, >97% by SDS-PAGE)
Product Number L2898 (aseptically filled, lyophilized powder, min. 500 units/mg protein)
Product number L9043 (lyophilized powder min 3000 unit/mg solid protein)
- Lysostaphin is a zinc endopeptidase with a molecular weight of approximately 25 kDa. Because lysostaphin cleaves the polyglycine cross-links in the peptidoglycan layer of the cell wall of Staphylococcus species it has been found useful for cell lysis and also as a potential anti-microbial therapeutic.
- pH Optimum for activity: pH ~ 7.5
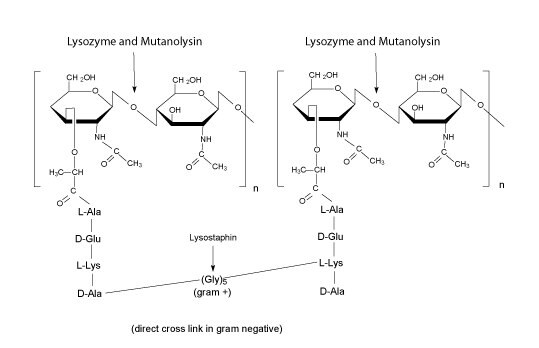
Figure 2.Lysozyme and Mutanolysin
Lysozyme from Chicken Egg White
Product Number L6876 (dialyzed, lyophilized, approx 50,000 un/mg)
Product Number L7651 (molecular biology grade, lyophilized, approx 50,000 un/mg)
Product Number L/CN/en7001 (lyophilized, approx 50,000 un/mg)
Product Number L7773 (aseptically filled, lyophilized, approx 50,000 un/mg)
Product Number L2879 (chloride, lyophilized, approx 60,000 un/mg)
Human Lysozyme
Product Number L1667 (recombinant expressed in rice)
Product Number L6394 (from human milk)
Product Number L8402 (from human neutrophils)
- Lysozyme is a single chain protein with a MW of 14.3 kD. It hydrolyzes β(1-4)-linkages between N-acetylmuraminic acid and N-acetyl-D-glucosamine residues in peptidoglycans and between N-acetyl-D-glucosamine residues in chitodextrin. Gram-positive cells are quite susceptible to this hydrolysis as their cell walls have a high proportion of peptidoglycan. Gram-negative bacteria are less susceptible due to the presence of an outer membrane and a lower proportion of peptidoglycan. However, these cells may be hydrolyzed more easily in the presence of EDTA that chelates metal ions in the outer bacterial membrane.
- Optimal pH: The activity of lysozyme is a function of both pH and ionic strength. The enzyme is active over a broad pH range (6.0 to 9.0). At pH 6.2, maximal activity is observed over a wider range of ionic strengths (0.02 to 0.100 M) than at pH 9.2 (0.01 to 0.06 M).
- For the lysis procedure, use a freshly prepared solution of 500,000 units/ml in 10 mM Tris-HCl, pH 8.0. The product is also soluble in water (10 mg/ml) yielding a clear to slightly hazy colorless solution. Aqueous solutions should be stable for at least one month when stored between 2–8 °C refrigerated. 25 µl Of this stock solution will typically lyse E. coli from >1ml of culture media cell pellet resuspended in 350 µl 10 mM Tris-HCl, pH 8.0, with 0.1 M NaCl, 1 mM EDTA, and 5% [w/v] TRITON. X-100. Typical incubation conditions for lysis are 30 min at 37 °C.
Achromopeptidase
Product Number A3422 (partially purified powder, 20,000-40,000 units/mg solid)
Product Number A3547 (crude powder, 800-3,200 units/mg solid)
Product Number A7550 (crude powder, 300-600 units/mg solid)
- Achromopeptidase is a lysyl endopeptidase with a MW of ~27kD. It is useful for lysis of Gram-positive bacteria that are resistant to lysozyme.
- pH Optimum for activity: pH ~ 8.5 - 9
- Approximately 500-1,500 un/ml achromopeptidase can be used to lyse cells at a density of A600=0.6 over 2 hours at 37 °C
Mutanolysin from Streptomyces globisporus
Product Number SAE0092 (lyophilized, min 4000un/mg protein)
Product Number M9901 (lyophilized, min 4000un/mg protein)
Product Number M4782 (Aseptically filled, lyophilized, min 4000un/mg protein)
- Mutanolysin provides gentle cell lysis for the isolation of easily degradable biomolecules and RNA from bacteria. It has been used in the formation of spheroplasts for isolation of DNA.
- Mutanolysin is a 23kD N-Acetyl Muramidase, like lysozyme, is a muralytic enzyme that cleaves the N-acetylmuramyl-β(1-4)-N-acetylglucosamine linkage of the bacterial cell wall polymer peptidoglycan-polysaccharide. Its carboxy terminal moieties are involved in the recognition and binding of unique cell wall polymers. Mutanolysin lyses Listeria and other gram positive bacteria such as Lactobacillus and Lactococcus.
α-Hemolysin from Staphylococcus aureus
Product Number H9395
- α-Hemolysin is an extracellular protein secreted by most strains of pathogenic Staphylococcus aureus.
- The mature protein contains 293 residues with a molecular weight of 33 kD. It is composed of 65% β-sheets and 10% α-helical structures.
- α-Hemolysin is secreted as a water-soluble monomer which initially binds and incorporates into the target cell membrane. The monomers assemble to form a heptametric pore The pore allows rapid efflux of K+ and influx of NA+ and Ca+. Osmotic swelling of erythrocytes finally results in a rupture.
Figure 3.Staphylococcus aureus
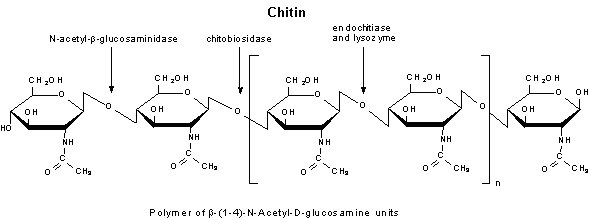
Figure 4.Chitinase
- Chitinase is a 30 kDa (approx.) extracellular enzyme complex that degrades chitin.
- Chitin is degraded to N-acetyl-D-glucosamine in 2 enzymatic reactions. Firstly, chitobiose units are removed from chitin by chitodextrinase-chitinase, a poly(1,4-β-[2-acetamido-2-deoxy-D-glucoside])-glycanohydrolase. The second reaction involves N-acetyl-glucosaminidase-chitobiase, which cleaves the disaccharide to its monomer subunits of N-acetyl-D-glucosamine.
- The enzyme may be classified into endo- and exochitinase. The endochitinase activity involves random cleavage at internal points in the chitin chain. The exochitinase activity consists of a progressive action which starts at the non-reducing end of chitin and releases chitobiose or N-acetyl-glucosamine units.
The microbiome – sequencing and phylogeny
The study of microbial communities has recently been revolutionized by the widespread adoption of techniques such as 16S rRNA gene sequencing and metagenomics. Since DNA contamination during microbiome sample preparation is a major challenge of these sequence-based approaches, DNA extraction reagents free of additional DNA contaminants are essential.
Another major challenge in processing microbiome samples is that microbes are difficult to disrupt - the cell walls can form capsules or resistant spores when processed. DNA can be extracted from the microbes by using lysing enzymes to induce partial spheroplast formation. These spheroplasts are subsequently lysed to release DNA.
DNA-free lytic enzymes for studying the microbiome
We offer a growing list of microbial DNA-free lytic enzymes and enzyme mixes that will help improve lysis of microbiome samples without random DNA contamination.
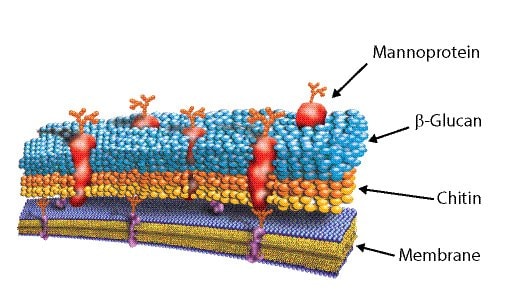
Figure 4.Yeast cell wall
In Yeast, the cell wall comprises ~30 % of the dry weight of the cell. The yeast cell wall is made of ~25% helical β(1-3) and β(1-6)-D-glucans and ~25% oligo-mannans, ~20 % protein, ~10% lipids, and some chitin. The protein component exists predominantly as a mannoprotein complex. Covalent linkages are reported to exist as β(1-4)-linkages between the reducing ends of chitin and the nonreducing end of b(1-3)-glucans as well as among glycoproteins, β(1-6)-glucans, and β(1-3)-glucans.
Enzymes That Can Break Down the Yeast Cell Wall
Lysing Enzymes from Rhizoctonia solani (Kitalase)
Product Number L8757 (powder)
- Main enzymatic acitivity is β-1,3-glucanase, also reported to contain protease, pectinase and amylase activities.
Lyticase from Arthrobacter luteus
Product Number L2524 (powder, min 2,000 un/mg protein)
Product Number L4025 (powder, min 200 un/mg solid)
Product Number L5263 (Zymolyase® 100Tpowder, min 3,000 un/mg protein)
- Contains β-1,3-glucan laminaripentaohydrolase with additional β-1,3-glucanase, protease and mannanase activities. Trace activities may include xylanase, amylase and phosphatase.
- Reported to be useful for lysis of Ashbya, Candida, Debaryomyces, Eremothecium, Endomyces, Hansenula, Hanseniaspora, Kloeckera, Kluyveromyces, Lipomyces, Metschikowia, Pichia, Pullularia, Torulopsis, Saccharomyces, Saccharomycopsis, Saccharomycodes, and Schwanniomyces.
Toxins for Eukaryotic Cells
A toxin is a poisonous small molecule, peptide, or protein produced within living cells. Some toxins interact with molecules on surface of a cell in a way that enhances the entry into the interior of the cell. These toxins are useful tools for researchers that want to disrupt the cell.
Making Eukaryotic Cells Permeable- with a toxin
α-Hemolysin from Staphylococcus aureus
Product Number H9395 (lyophilized powder, ≥10,000 units/mg protein)
- α-Hemolysin is a 33 kD extracellular protein secreted by most strains of pathogenic Staphylococcus aureus.
- It is selectively hemolytic and has a marked preference for rabbit red blood cells.
- It induces dermonecrosis, muscle paralysis, and it is lethal for laboratory animals.
- The toxin must be in the monomeric form to initially bind to a membrane. Specific receptors are not required for binding. Upon binding to biological membranes and/or artificial membranes, self-oligomerization occurs, resulting in ring structures (hexameric aggregates) believed to be transmembrane pores, which are permeable to ions and small metabolites.
- It is thought that α-hemolysin stimulates cellular phospholipases and induces a Ca2+ influx that can result in membrane disruption, leakage of cytoplasmic components, impaired membrane permeability, and osmotic lysis of the cells.
Streptolysin O from Streptococcus pyogenes
Product Number SAE0089
Product Number S5265
Product Number S0149
- Streptolysin O is a toxin that is a single polypeptide chain with a molecular weight of 62 kDa.
- Streptolysin O binds to membrane cholesterol and oligomerizes to create a ring structure that consists of 45 to 50 units. The ring structure inserts into the membrane to make a large pore (25 to 30 nm), which DNA, RNA, peptides and proteins may pass.
Figure 5.Streptococcus Pyogenes
Tetanolysin from Clostridium tetani
Product Number T5319 (lyophilized powder)
- Cholesterol-binding toxin used to permeabilize cellular membranes to enhance the entry of macromolecules into the interior of the cell. Pores induced reported to be in the range of 20-50 nm.
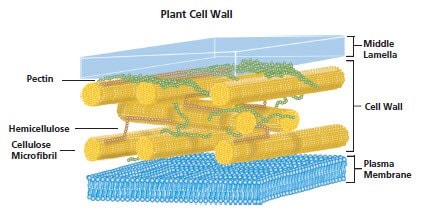
Figure 6.Plant Cell Wall
Plant cells are surrounded by a rigid, semi-permeable cell wall. The cell wall is comprised mainly polysaccharides with some proteins and lipids. The three main polysaccharide components of the cell wall are cellulose, an unbranched polymers of β-(1-4)-D-glycopyranosyl units associated in microfibril bundles. The microfibrils are cross-linked by hemicellulose (a branched polymer of β-(1-4)-D-xylopyranosyl units). This cross-linked structure is embedded in a matrix of pectin (primarily containing an α-(1-4) polygalacturonic acid backbone which can be randomly acetylated and methylated.
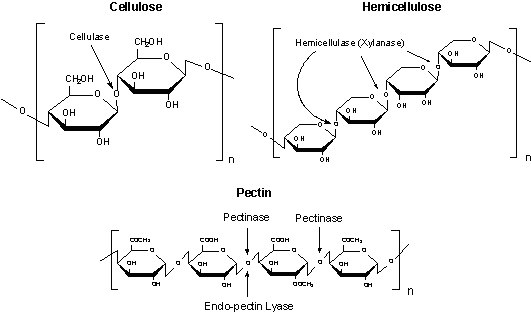
Figure 7.Cellulose, Hemicellulose and Pectin structures
Cellulase
Cellulase preparations are typically mixtures of enzymes containing high cellulase activity with some hemicellulase activity. These enzyme mixtures are capable of degrading cellulose, mannans, xylans, galactomannans, pectins and other polysaccharides.
from Aspergillus species
Product Number C2605 (Novozymes Carezyme 1000 L)
Product Number V2010 (Novozymes Viscozyme L)
from Aspergillus niger
Product Number C1184 (powder, min. 0.3 un/mg)
from Basidiomycetes sp
Product Number D9515 (Driselase, powder)
from Trichoderma reesei
Product Number C8546 (powder, min. 1.0 un/mg)
Product Number C2730 (Novozymes Celluclast 1.5 L)
from Trichoderma viride
Product Number C0615 (Yakult Onozuka RS)
Product Number C1794 (cell culture tested, powder, min. 3.0-10 un/mg)
Product Number C9422 (powder, min. 3.0-10 un/mg)
Pectinase and Pectolyase
Pectinase catalyzes the random hydrolysis of 1-4-α-D-galactosiduronic linkages in pectin and other galacturonans. Pectolyase catalyzes the eliminative cleavage of (1-4)-α-D-galacturonan methyl ester to give oligosaccharides with 4-deoxy-6-O-methyl- α-D-galact-4-enuronosyl groups at their non-reducing ends.
Pectinase
from Aspergillus aculeatus
Product Number P2611 (Novozymes Pectinex Ultra SPL)
from Aspergillus niger
Product Number P2736 (Novozymes Pectinex 3XL)
Product Number P4716 (aqueous glycerol solution, min. 5 un/mg protein)
Product Number P0690 (plant cell culture tested)
from Rhizopus species
Product Number P2401 (Macerozyme R10 Crude Powder 400-800 un/g)
Product Number P4300 (plant cell culture tested)
Pectolyase
from Aspergillus japonicus
Product Number P3026 (lyophilized, min. 0.3 un/mg)
Product Number P5431 (lyophilized, min. 2 un/mg)
Product Number P5936 (plant cell culture tested)
References
To continue reading please sign in or create an account.
Don't Have An Account?